PADI3 induces cell cycle arrest via the Sirt2/AKT/p21 pathway and acts as a tumor suppressor gene in colon cancer
Xiaotian Chang, Zhengbin Chai, Jiaorui Zou, Hongxing Wang, Yao Wang, Yabing Zheng, Hui Wu,Chunyan Liu
1Medical Research Center, The First Affiliated Hospital of Shandong First Medical University, Jinan 250014, China; 2Medical Research Center of the Hospital Affiliated to Qingdao University, Qingdao 266000, China; 3 Department of Laboratory Medicine, Jinan Infectious Disease Hospital, Shandong University, Jinan 250021, China
ABSTRACT Objective: As a member of the peptidyl arginine deiminase (PAD) family, PADI3 is weakly expressed in colon cancer tissues and highly expressed in adjacent colon cancer tissues. However, the role of PADI3 in colon cancer is unclear. In this study, we investigated the function and molecular mechanism of PADI3 in colon cancer tumorigenesis.Methods: Western blot and real-time PCR were used to detect the expression levels of several genes. CCK-8, flow cytometry(FCM) and colony formation assays were used to examine cell proliferation, the cell cycle and colony formation ability. RNAsequencing analysis was used to study the molecular mechanism of PADI3 in tumorigenesis. A truncation mutation experiment was performed to determine the key functional domain of PADI3.Results: PADI3 overexpression inhibited cell proliferation and colony formation and led to G1 phase arrest in both HCT116(originating from primary colon cancer) and LoVo (originating from metastatic tumor nodules of colon cancer) cells. PADI3-expressing HCT116 cells had a lower tumor formation rate and produced smaller tumors than control cells. PADI3 significantly decreased Sirtuin2 (Sirt2) and Snail expression and AKT phosphorylation and increased p21 expression, and Sirt2 overexpression partly reversed the effects induced by PADI3 overexpression. Immunocytochemistry showed that PADI3 is mainly localized in the cytoplasm. Truncation mutation experiments showed that the C-domain is the key domain involved in the antitumor activity of PADI3.Conclusions: PADI3 suppresses Snail expression and AKT phosphorylation and promotes p21 expression by downregulating Sirt2 expression in the cytoplasm, and the C-domain is the key domain for its antitumor activity.
KEYWORDS PADI3; Sirt2; colon cancer; cell cycle; C-domain
Introduction
Colorectal cancer (CRC) is a disease with high mortality worldwide1,2, but the pathogenesis of CRC remains unclear3.Surgical interventions, chemotherapy and radiotherapy are the major treatments for CRC in the clinic. Despite these treatments, the poor prognosis associated with metastasis and recurrence postsurgery has not improved4,5. Further study of the molecular mechanism of colon cancer, the identification of colon cancer-suppressing genes, and methods to improve the survival rate of patients are issues that urgently need to be addressed.
Citrullination, a posttranslational modification, is catalyzed by a group of enzymes called peptidyl arginine deiminases (PADs). PADs are reported to be involved in various biological processes in numerous human diseases,such as rheumatoid arthritis6, Alzheimer’s disease7, ulcerative colitis8, and cancers9,10. The PAD family comprises five members: PADI1, PADI2, PADI3, PADI4 and PADI6. Many reports have shown that PADI1, PADI2 and PADI4 are involved in the tumorigenesis of various cancers10-13, and PADI6 is involved in early embryonic development14. PADI3 has been reported to be expressed in human keratinocytes and plays an important role in uncombable hair syndrome15,16. However, the role of PADI3 in tumorigenesis is poorly understood.
In the present study, we found that PADI3 is highly expressed in adjacent colon tissues, but only a small amount of PADI3 expression can be detected in colon cancer tissues.We therefore further investigated the function of PADI3 in the tumorigenesis of colon cancer. In vitro, the overexpression of PADI3 inhibited cell proliferation by inducing G1 phase arrest. In vivo, PADI3-expressing HCT116 cells showed a lower tumor formation ratio and produced smaller tumors compared with the control group.We speculate that PADI3 plays an antitumor role in colon cancer. Further study showed that PADI3 can significantly suppress Sirt2 expression at both the transcriptional and translational levels. However, the molecular mechanism of this process is still poorly understood.
Sirt2 belongs to the Sirtuin family, which possesses NAD+-dependent protein deacetylase activity17. Sirt2 participates in a variety of cellular processes, including the cell cycle, the mitotic checkpoint, autophagy, apoptosis, migration and invasion18-20. Sirt2 is highly expressed in multiple tumors21-23. Increasing evidence has demonstrated that Sirt2 plays an important role in tumorigenesis, but the function of Sirt2 in tumorigenesis is still under debate17,21,24-26. The opposing functions of Sirt2 in tumorigenesis may be dependent on the tissue distribution of it.
Inhibitors of Sirt2, such as TM, AK1, AGK-2, AC-93253,AEM1, AEM2, and Tenovin-D3, have been proven to suppress tumorigenesis in a variety of tumors17,23,27-32.According to a previous study, effective Sirt2 inhibitors are considered to be the most promising cancer treatment. In colon cancer, suppressing Sirt2 expression can induce cell cycle arrest17, promote cell apoptosis33, and inhibit tumor angiogenesis34. Suppressing Sirt2 expression may be a wise choice in colon cancer therapy. Based on the above results,we speculate that PADI3 has antitumor activity as a Sirt2 inhibitor, but the molecular mechanism is still unclear.
Further study showed that PADI3 can significantly decrease Snail expression and AKT phosphorylation while increasing p21 expression via the downregulation of Sirt2 expression. Truncation mutation and immunocytochemistry showed that PADI3 exerts its antitumor activity mainly in the cytoplasm and that the C-domain is the key functional domain of PADI3.
These findings indicate that PADI3 exerts its antitumor activity by suppressing Sirt2 expression in the cytoplasm and that the C-domain is essential for PADI3 to exert its antitumor activity.
Materials and methods
Animals
BALB/c nude mice were housed in an Association for Assessment and Accreditation of Laboratory Animal Care(AAALAC)-accredited facility. The study was approved by the Ethics Committee of Shandong Province Qianfoshan Hospital, Jinan, China (Approval No. S0087).
Primers and antibodies
Full-length of PADI3 was amplified using PCR with the primers PADI3-EcoRI-Fex and PADI3-AscI-Rex, the Ndomain was amplified with the primers PADI3-N-OEEcoRI-Fex and PADI3-N-OE-AscI-Rex, the M-domain was amplified with the primers PADI3-M-OE-EcoRI-Fex and PADI3-M-OE-AscI-Rex, the C-domain was amplified with the primers PADI3-C-OE-EcoRI-Fex and PADI3-C-OEAscI-Rex, the NM-domain was amplified with the primers PADI3-N-OE-EcoRI-Fex and PADI3-M-OE-AscI-Rex, and the MC-domain was amplified with the primers PADI3-M-OEEcoRI-Fex and PADI3-C-OE-AscI-Rex. The primer sequences were designed as shown in Supplementary Table S1.
The following primary antibodies used in this study were commercially obtained: anti-PADI3 (Abcam, ab172959),anti-GAPDH (Abcam, ab181603), anti-AKT (Santa Cruz Biotechnology, sc5298), anti-Sirt2 (Santa Cruz Biotechnology, sc28298), anti-p21 (Santa Cruz Biotechnology, sc6246), anti-phosphorylated AKT (p-AKT)(CST, #4060), anti-Snail (CST, #3879), anti-His-tag (CST,12698S), anti-histone 3 (Affinity, AF0863) and anti-red fluorescent protein (RFP) (Abcam, ab28664). The following secondary antibodies were used in this study: goat anti-rabbit(Affinity, #s0001), goat anti-mouse (Affinity, #s0002) and donkey anti-goat (Abcam, ab6881).
Quantitative real-time-PCR analysis
The total RNA of cells or tissues was extracted using Trizol reagent (Life Technologies, USA). The extracted RNA was reverse-transcribed into first-strand cDNA in a final volume of 10 μ L using an RNA PCR Kit (Toyobo, Japan). The reverse-transcribed first-strand cDNA was used as the template for real-time PCR with the forward primer PADI3-QF and the reverse primer PADI3-QR; the primer sequences are shown in Supplementary Table S1. The conditions of real-time PCR were as follows: 10 s at 95°C; 45 cycles of 5 s at 60°C and 10 s at 72°C; and 30 s at 65°C. This experiment was performed in triplicate. Real-time PCR was performed using a 10 μL total volume that contained 1 μL of cDNA, 2 μL of ddH2O, 5 μ L of SYBR Green Real-time PCR Master Mix(Toyobo, Japan), 1 μL of forward primer and 1 μL of reverse primer. GAPDH was used for quantity and quality control using the forward primer of human GAPDH-QF and the reverse primer of human GAPDH-QR. The primer sequences are shown in Supplementary Table S1. Data were analyzed using the formula R = 2-[ΔCtsample-ΔCtcontrol], where R is the relative expression level, ΔCt sample is the difference between the Ct of the gene and the average GAPDH in the experiment sample, and ΔCt control is the difference between the Ct of the gene and the average GAPDH in the control sample.
Western blot analysis
One hundred micrograms of tumor tissue was homogenized in 600 μL of Cell Lysis Solution (Sigma-Aldrich, USA) and centrifuged at 12,000 rpm for 20 min at 4°C. The supernatant was collected after centrifugation, and the protein concentrations were determined using the Bradford Protein Assay Kit (Beyotime, China). Total protein was extracted and then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene fluoride membrane (Millipore, USA).Western blot analysis was conducted using the anti-PADI3 antibody. The acquisition of enhanced chemiluminescence(ECL) images was carried out with a Typhoon Trio system(GE Healthcare, USA).
Construction of PADI3-expressing plasmids
The full open reading frame (ORF) of PADI3 is 1,995 bp and encodes 664 amino acid residues. PADI3 cDNA was synthesized by YouBio (China). PADI3 was amplified by PCR using the PADI3-EcoRI-Fex and PADI3-AscI-Rex primers. The PCR products were inserted into the pCDNA3.1-RFP expression vector, which contains a His-tag and is resistant to ampicillin. RFP was selected as the reporter gene, which was inserted into the multiple cloning site(MCS) using the restriction enzymes Kpn I and NotI.Recombinant pCDNA3.1-PADI3-RFP plasmids were purified using a GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific, USA) and stored at —80°C.
Cell proliferation assay
Cell proliferation assays were performed using a CCK-8 kit to determine the effect of PADI3 on HCT116 and LoVo cell proliferation. Recombinant pCDNA3.1-PADI3-RFP plasmids were transfected into HCT116 or LoVo cells using PolyJet DNA In Vitro Transfection Reagent (SignaGen, USA)according to the manufacturer’s instructions. After culturing for 48 h at 37°C with 5% CO2, CCK-8 solution (Toyobo,Japan) at a concentration of 10 μL/well was added to the cell culture medium, and culturing was continued for another 2 h.The absorbance was measured at 450 nm with a spectrophotometer (SpectraMax 190, Molecular Devices,USA). Graphs showing growth were generated from the average values of five wells in each group.
Cell colony formation assay
HCT116 cells were transfected with pCDNA3.1-PADI3-RFP plasmids to overexpress PADI3, and HCT116 cells transfected with pCDNA3.1-RFP plasmids were used as controls. Following transfection for 48 h, the cells were harvested, and 1000 cells in 3 mL of complete Dulbecco’s Modified Eagle Medium (DMEM) were seeded into a 6-cmdiameter Petri dish. After 10 days of culturing, colonies were fixed with methanol and stained with 0.25% crystal violet.Colony numbers and sizes from three independent experiments were analyzed.
Cell cycle measurements using flow cytometry(FCM) via propidium iodide (PI) staining methods
PADI3-expressing HCT116 cells were prepared by the transfection of p3×FLAG-CMV-PADI3-7.1 recombinant plasmids. Cells transfected with blank p3×FLAG-CMV-7.1 plasmids were used as the negative control. Following transfection for 60 h, the cells were harvested and washed 2 times with precooled phosphate-buffered saline (PBS) and then stored at —20°C with 70% ethanol overnight. A DNA Content Quantitation Assay (Cell Cycle) Kit (Solarbio,China) was used to analyze the cell cycle. The procedure was as follows: First, overnight-fixed cells were washed twice with 1 mL of precooled PBS, and after centrifugation at 1,000 rpm for 5 min at room temperature, the cells were resuspended in 100 μL of RNase A and incubated at 37°C for 30 min. Next,400 μL of PI solution was added to the cell-RNase A mixture and incubated at 4°C for 30 min in the dark. Finally, the cells were washed with precooled PBS and then analyzed using FCM.
PADI3 gene structure prediction analysis
The PADI3 sequence was obtained from GenBank(https://www.ncbi.nlm.nih.gov/genbank/). Sequence analysis was performed using BLASTX software (http://www.ncbi.nlm.nih.gov/). Gene translation and protein prediction were performed using ExPASy (http://www.au.expasy.org/).Domain prediction was performed using SMART software(http://smart.embl-heidelberg.de/).
Construction of plasmids expressing truncated PADI3
Five expression plasmids that contain different parts of the PADI3 gene-encoding region were reconstructed. The Ndomain (aa 1-113), M-domain (aa 115-273), C-domain (aa 283-661), NM-domain (aa 1-273) and MC-domain (aa 115-661) of the PADI3 gene were amplified by PCR using the primer pairs PADI3-N-OE-EcoRI-Fex and PADI3-N-OEAscI-Rex, PADI3-M-OE-EcoRI-Fex and PADI3-M-OE-AscIRex, PADI3-C-OE-EcoRI-Fex and PADI3-C-OE-AscI-Rex,PADI3-N-OE-EcoRI-Fex and PADI3-M-OE-AscI-Rex, and PADI3-M-OE-EcoRI-Fex and PADI3-C-OE-AscI-Rex,respectively. The primer sequences are shown in Supplementary Table S1. These PADI3 cDNA sequences were inserted into the MCS of pCDNA3.1-RFP expression vectors using the restriction enzymes EcoR I and Asc,respectively. The truncated proteins were expressed in HCT116 cells. Cells transfected with pCDNA3.1-RFP plasmids were used as controls.
Immunocytochemistry (ICC)
PADI3-expressing cells were fixed in 4% paraformaldehyde for 10 min at room temperature, washed twice with PBS and permeabilized with 0.2% Triton X-100 in PBS at 37°C for 30 min. The nuclei were stained with 4', 6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, USA) for 10 min at room temperature. After washing twice with PBS, the cells were observed using an Olympus FSX100 fluorescence microscope (Olympus, Japan).
Establishment of tumor-bearing mice with HCT116 cells
The full coding sequence of PADI3 was inserted into pHBLV-CMVIE-T2A-Puro vectors and coated with lentiviral(Hanbio, China). The lentiviral coated pHBLV-CMVIEPADI3- T2A-Puro vectors were transfected into HCT116 cells and screened with 2 μ g/mL puromycin. pHBLVCMVIE-RFP-T2A-Puro-transfected HCT116 cells were used as controls. Thirty 6-week-old BALB/c nude mice (Vital River, China) were used to establish a tumor-bearing mouse model. Two hundred microliters of PADI3-expressing HCT116 cells (107/mL in PBS) were injected into the right dorsal flank of each mouse, and 200 μL of cells transfected with the blank vectors were injected into the left dorsal flank of each mouse. Tumors were dissected at 42 days after cell injection; tumor size and weight were measured by routine methods.
RNA-sequencing analysis
HCT116 cells were transfected with pCDNA3.1-PADI3-RFP plasmids, and cells transfected with pCDNA3.1-RFP plasmids were used as controls. After 60 h of culture, the cells were harvested, and total RNA was extracted using a mirVanaTMmiRNA Isolation Kit (Thermo Fisher Scientific,USA). The quality of the total RNA was evaluated using an Agilent 2100 Bioanalyzer (Agilent, USA), and RNAsequencing was performed using an Illumina HiSeqTM 2500 system. Three independent experiments were performed to verify the results. Genes showing a ≥ 3-fold change in their expression level were selected for further study.
Cytoplasmic and nuclear protein extraction
A Nuclear and Cytoplasmic Protein Extraction Kit(Beyotime, China) was used to extract nuclear and cytoplasmic proteins. Cultured cells were washed with precooled PBS, harvested with a scraper, and collected by centrifugation. The cell pellet was resuspended with 200 μL of PMSF-supplemented Cytoplasmic Protein Extraction Reagent A, vortexed for 5 s at maximum speed, and incubated in an ice bath for 10—15 min. Following the addition of 10 μL of Cell Paste Protein Extraction Reagent B,the cell lysates were vortexed for 5 s at maximum speed and centrifuged at 16,000× g and at 4°C for 5 min. The extracted cytoplasmic proteins were in the supernatant and collected.The remaining pellet was added to 50 μL of nuclear protein extraction reagent supplemented with PMSF, vortexed for 15—30 s at maximum speed, incubated in an ice bath briefly and vortexed again for 15—30 s every 1—2 min for a total of 30 min. Finally, the solution was centrifuged at 16,000× g and at 4°C for 10 min. The nuclear proteins were collected from the supernatant.
Construction of Sirt2-expressing plasmids
The full coding sequence of Sirt2 isoform 1 (Sirt2-iso1) is 1,170 bp and encodes 389 amino acid residues; the full coding sequence of Sirt2 isoform 2 (Sirt2-iso2) is 1,059 bp and encodes 352 amino acid residues. The cDNA sequences were synthesized and sequenced by Generay (Shanghai,China). Sirt2-iso1 was amplified using Sirt2-iso1-Fex and Sirt2-iso1-Rex primers. Sirt2-iso2 was amplified using Sirt2-iso2-Fex and Sirt2-iso2-Rex primers. The primer sequences are shown in Supplementary Table S1. The PCR products were inserted into pCDNA3.1-GFP expression vectors, which contain a His-tag and a GFP reporter gene. For rescue experiments, cultured HCT116 cells were transfected with pCDNA3.1-PADI3-RFP and cultured for 24 h to express the PADI3 protein. PADI3-overexpressing HCT116 cells were then transfected with the pCDNA3.1-Sirt2-iso1-GFP or pCDNA3.1-Sirt2-iso2-GFP plasmid and continually cultured for 48 h. Then, the cells were harvested and further examined.
Statistical analysis
Data were analyzed by a two-tailed Student’s t test.Differences were considered to be statistically significant at P < 0.05. To verify the results, each experiment was performed with three samples in triplicate.
Results
Expression pattern of PADI3 in colon cancer tissues
To study the function of PADI3 in tumorigenesis, the expression profile of PADI3 in various cancers, including breast cancer, colon cancer, esophageal cancer, gastric cancer,hepatocellular carcinoma and lung cancer, were detected using real-time PCR. Results showed that only in colon cancer was PADI3 significantly different between cancer tissues and corresponding adjacent tissues (Supplementary Figure S1).
The expression pattern of PADI3 in colon cancer tissue and corresponding adjacent tissue samples obtained from 20 different patients was investigated using western blot analysis and qRT-PCR to verify the above results. There was only a little or no expression of PADI3 in the colon cancer tumor tissues; however, a high level of PADI3, with a molecular weight of 75 kDa, was detected in the corresponding adjacent tissues (Figure 1).
Effect of PADI3 expression on cell proliferation and colony formation

Figure 1 PADI3 expression profile determination using Western blot analysis and qRT-PCR. (A) Western blot analysis was used to measure the expression level of PADI3 in colon cancer tissue samples and corresponding adjacent tissue samples at the translational level. These paired tissue samples were obtained from 20 different patients; GAPDH was selected as the internal control. (B) Statistical analysis of Western blot. (C) qRT-PCR was used to measure the expression level of PADI3 in the colon cancer tissue samples and corresponding adjacent tissue samples at the transcriptional level. T: tumor tissues; N: corresponding adjacent tissues. * indicates P < 0.05, and ** indicates P < 0.01 for three independent experiments analyzed by Student’s t-test.

Figure 2 Effects of PADI3 on HCT116 and LoVo cells. RFP-expressing cells were used as controls. (A) Weak PADI3 expression was detected in HCT116 and LoVo cells. (B) Western blot analysis was used to measure PADI3 and RFP expression in both LoVo and HCT116 cells. (C, D) A CCK-8 assay measured the proliferation of HCT116 and LoVo cells. (E) The colony formation ability of HCT116 cells was measured and statistically analyzed using a colony formation assay following 14 days of culture. (F) The colony formation ability of LoVo cells was measured and statistically analyzed using a colony formation assay following 14 days of culture. * indicates P < 0.05 for three independent experiments analyzed by Student’s t-test.
HCT116 and LoVo cells were transfected with the pCDNA3.1-PADI3-RFP plasmid, respectively. Western blot analysis detected weak PADI3 expression in the parental HCT116 and LoVo cells (Figure 2A). A high level of PADI3,with a molecular weight of 106 kDa (including RFP, 25 kDa;PADI3, 75 kDa; and His-tag, 6 kDa), was detected in the HCT116 and LoVo cells using the anti-His antibody. The RFP immunosignal, with a molecular weight of 31 kDa(including RFP, 25 kDa, and His-tag, 6 kDa), was also detected in the two cell lines following transfection with the pCDNA3.1-RFP plasmid without PADI3 expression. These results demonstrated the production of PADI3-RFP protein in HCT116 and LoVo cells following transfection (Figure 2B).These findings indicate that there is only weak PADI3 expression in untransfected HCT116 and LoVo cells and demonstrate the successful transfection and overexpression of PADI3 in the two cell lines.
CCK-8 assays were performed to measure the proliferation of HCT116 and LoVo cells after PADI3 overexpression. The analysis detected a significant decrease in cell proliferation in the PADI3-expressing HCT116 (P = 0.004,8) and LoVo (P =0.007,1) cells compared with the corresponding control cells(Figure 2C, D).
Colony formation assays were performed to determine the antitumor role of PADI3 after transfection into HCT116 and LoVo cells. The results showed that the overexpression of PADI3 significantly reduced the number and size of colonies in both HCT116 and LoVo cells compared with the control cells (P < 0.001) (Figure 2E, F).
Function of PADI3 in vivo
Tumor-bearing mice were generated using HCT116 cells that were transfected with lentiviral packaging pHBLV-CMVIEPADI3-T2A-Puro plasmids. HCT116 cells that were transfected with lentiviral packaging pHBLV-CMVIE-RFPT2A-Puro plasmids were used as the nonspecific control.Western blot analysis confirmed PADI3 expression in the transfected HCT116 cells (Figure 3A). At 42 days after cell injection, the tumor-bearing mice were sacrificed. All 9 mice showed obvious tumor growth on the left side of their body following the injection of HCT116 cells that were transfected with RFP-expressing lentiviruses. In contrast, 6 of these mice showed minimal tumor growth on the right side of the body following the injection of cells that were transfected with PADI3-expressing lentiviruses, while the other 3 mice did not show obvious tumor growth on the right side (Figure 3B,3C). The tumors on the right side were much smaller and the tumor formation ratio was much lower than those on the left side (Figure 3D, 3E ). These results indicate that the overexpression of PADI3 significantly suppresses tumor growth and tumorigenesis in vivo.
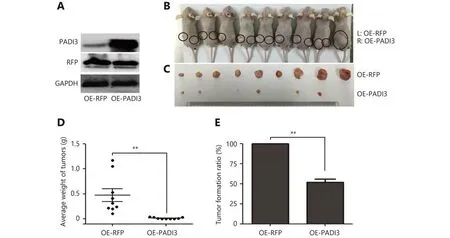
Figure 3 Effects of PADI3 in vivo. (A) Western blot analysis was used to measure PADI3 expression. (B) PADI3-expressing HCT116 cells were injected into the left dorsal flank of BALB/c nude mice, and RFP-expressing HCT116 cells were injected into the right dorsal flank.(C) Tumors were dissected 42 days after cell injection. (D) Tumor weight was measured and statistically analyzed. (E) The tumor formation ratio was statistically analyzed. * indicates P < 0.05.
Function of PADI3 in the cell cycle
Cell cycle progression was measured using the DNA Content Quantitation Assay (Cell Cycle) via FCM. The proportion of cells in G1 phase arrest was significantly elevated in HCT116 cells transfected with PADI3-FLAG-expressing plasmids compared with control cells transfected with FLAGexpressing plasmids (P = 0.005,7) (Figure 4A-4C). In LoVo cells, similar results were also observed; G1 phase arrest was significantly elevated in LoVo cells transfected with PADI3-FLAG-expressing plasmids compared with control cells transfected with FLAG-expressing plasmids (P = 0.002,9)(Figure 4D-4F). These results indicate that PADI3 suppresses cell proliferation by inducing G1 phase arrest in both HCT116 and LoVo cells.
Downstream regulatory mechanism of PADI3
RNA-sequencing assays were performed using HCT116 cells transfected with pCDNA3.1-PADI3-RFP plasmids, and HCT116 cells transfected with pCDNA3.1-RFP plasmids were used as controls. The RNA-sequencing analysis demonstrated that the overexpression of PADI3 in HCT116 cells led to a significant decrease in the expression of Sirt2 and U2 small nuclear RNA auxiliary factor 1 (U2AF1) and a marked increase in the expression of spondin 1 (SPON1),growth arrest and DNA-damage-inducible 45G (GADD45G),myostatin (MSTN), ribosomal protein S6 kinase polypeptide 1 (RPS6KA1), protein tyrosine phosphatase non-receptor type 22 (PTPN22) and type II iodothyronine deiodinase(DIO2) (Figure 5A).
To further investigate the downstream regulatory pathway of PADI3 during tumorigenesis, the expression of AKT,phosphorylated AKT (p-AKT), Snail and p21 in HCT116 cells was also measured using Western blot analysis. PADI3-overexpressing HCT116 cells exhibited significantly suppressed Sirt2, p-AKT and Snail expression and increased p21 expression compared with the control groups (Figure 5B).Sirt2 had two immunosignals with molecular weights of 40 kDa and 43 kDa, which most likely correspond to Sirt2-isoform 1 (Sirt2-iso1) and Sirt2-isoform 2 (Sirt2-iso2), in the HCT116 cells overexpressing PADI3.
To verify the relationship between PADI3 and Sirt2 in vivo,the expression patterns of PADI3 and Sirt2 in colon cancer tissues and corresponding adjacent tissue samples obtained from 6 different patients were investigated using Western blot analysis. The results showed that the expression patterns of PADI3 and Sirt2 were negatively correlated (Figure 5C, 5D).

Figure 4 Analysis of the roles of PADI3 in the cell cycle using FCM. (A) FLAG-expressing HCT116 cells were used as the negative control group. (B) PADI3-FLAG-expressing HCT116 cells were used as the experimental group. (C) Statistical analysis of A and B. (D) FLAGexpressing LoVo cells were used as the negative control group. (E) PADI3-FLAG-expressing LoVo cells were used as the experimental group.(F) Statistical analysis of D and E.

Figure 5 RNA-sequencing analysis of PADI3-expressing HCT116 cells. (A) Heat map from the RNA-sequencing analysis. (B) Expression of phosphorylated AKT (p-AKT), AKT, Sirt2, Snail and p21 in OE-PADI3 HCT116 cells, OE-RFP HCT116 cells is selected as the control group. (C)Western blot analysis was used to measure the expression level of Sirt2 and PADI3 in colon cancer tissues and corresponding tissues. (D)Statistical analysis of the expression level of Sirt2 and PADI3. * indicates P < 0.05 for three independent experiments analyzed by Student’s t-test.
Functional domain analysis of PADI3
The PADI3 gene structure was analyzed using SMART. The analysis indicated that PADI3 has 3 domains: the N-domain(aa 1-113, located in the N-terminal region), the M-domain(aa 115-273, located in the middle region, also named the PAD domain), and the C-domain (aa 283-661, located in the C-terminal region) (Figure 6A). The PADI3 fragments encoding the N-domain, M-domain, C-domain, NMdomain (containing both the M- and N-domains), and MCdomain (containing both the M- and C-domains) were individually inserted into pCDNA3.1-RFP plasmids. The recombinant plasmids containing full-length PADI3 or one of its five truncated forms were separately transfected into HCT116 cells to express the different domains of the enzyme.The cells transfected with full-length PADI3 were used as a positive control. Western blot analysis using the anti-His-tag antibody detected the expression of these truncated PADI3 gene fragments (RFP: 31 kDa, PADI3: 106 kDa, N-domain:44 kDa, M-domain: 49 kDa, C-domain: 74 kDa, NMdomain: 62 kDa, and MC-domain: 93 kDa) in HCT116 cells(Figure 6B). CCK-8 assays were performed to measure the proliferation of HCT116 cells transfected with the plasmids containing these PADI3 truncations. Compared with the HCT116 cells transfected with the pCDNA3.1-RFP plasmids,only the cells transfected with full-length PADI3, the PADI3 C-domain and the PADI3 MC-domain showed significantly inhibited cell proliferation. The cells transfected with the other domains showed similar results to those of the control cells (Figure 6C). These results suggest that the C-domain is essential for the antitumor roles of PADI3.
Subcellular locations of PADI3 and its truncated mutants in HCT116 cells
Immunocytochemistry was performed to determine the subcellular locations of the truncated PADI3 proteins in HCT116 cells. The immunosignals for the full-length protein and C-domain and MC-domain of PADI3 were mainly observed in the cytoplasm, whereas the signals for the Ndomain, M-domain and NM-domain of PADI3 were mainly detected in the nucleus (Figure 7A). Western blotting was also conducted to examine PADI3 expression in cytoplasmic and nuclear extracts. GAPDH and histone 3 (17 kDa) were used as the internal controls for the cytoplasm and nucleus,respectively. Full-length PADI3 was mainly detected in the cytoplasmic extracts; the N-domain, M-domain and NMdomain were mainly detected in the nucleus; and the Cdomain and MC-domain were detected in both the cytoplasmic and nuclear extracts (Figure 7B). The above results demonstrate that the C-domain of PADI3 anchors the enzyme in the cytoplasm and is critical to the role of PADI3 in inhibiting cell proliferation.

Figure 6 Functional analysis of different PADI3 domains. (A) Predicting the PADI3-domain structure using SMART. (B) Western blot analysis was used to detect the expression of different PADI3 domains in HCT116 cells. (C) CCK-8 assay was used to measure the cell proliferation activity of different PADI3 domains. * indicates P < 0.05 for three independent experiments analyzed by Student’s t-test.

Figure 7 Subcellular locations of the truncated forms of PADI3 in HCT116 cells. (A) Red fluorescence indicates RFP or PADI3-fused RFP,and blue indicates nuclei stained with DAPI. The merged image shows both RFP fluorescence and DAPI staining. The scale bar represents 20 μm. (B) Western blot analysis was used to detect truncated PADI3 in the cytoplasm and nucleus. GAPDH was used as the internal control for cytoplasmic expression, and histone 3 was used as the internal control for nuclear expression. Nu: nucleus, Cy: cytoplasm.
Molecular mechanism of PADI3 in inhibiting cell proliferation
A rescue experiment was performed, and pCDNA3.1-Sirt2-iso1-GFP (74 kDa) or pCDNA3.1-Sirt2-iso2-GFP (71 kDa)plasmids were transfected separately into PADI3-expressing HCT116 cells. HCT116 cells transfected with pCDNA3.1-RFP plasmids were used as negative controls, and HCT116 cells transfected with pCDNA3.1-PADI3-RFP plasmids were used as positive controls. The overexpression of Sirt2-iso1 or Sirt2-iso2 in PADI3-expressing HCT116 cells significantly increased AKT phosphorylation and Snail expression and suppressed p21 expression (Figure 8A). A CCK-8 assay showed that the overexpression of both Sirt2-iso1 (P = 0.035)and Sirt2-iso2 (P = 0.031) in the PADI3-expressing HCT116 cells significantly stimulated cell proliferation compared with the expression of PADI3 alone (Figure 8B). FCM with PI staining showed that the PADI3-expressing HCT116 cells overexpressing both Sirt2-iso1 (P = 0.0041) and Sirt2-iso2 (P =0.0023) exhibited significantly decreased G1 phase arrest compared with the control cells (Figure 8C, 8D). Colony formation assays showed that the overexpression of both Sirt2-iso1 (P = 0.0093) and Sirt2-iso2 (P = 0.0081) in PADI3-expressing HCT116 cells significantly stimulated colony formation (Figure 8E, 8F). These observations indicate that the overexpression of Sirt2-iso1/2 can significantly rescue the inhibitory effects of PADI3 on cell proliferation and colony formation and prevent the induction of G1 phase arrest in HCT116 cells.
Discussion
Citrullination is an important posttranslational modification process that is catalyzed by PADs. Earlier reports have shown that PADs are involved in various biological processes in numerous human diseases, and an increasing number of studies have found that PADs play important roles in tumorigenesis35-37. However, the role of PADI3 in tumorigenesis is not clear. In the present study, we found that PADI3 exerts its antitumor activity in colon cancer by regulating a variety of tumor-related genes and that the Cdomain is essential for PADI3 localization in the cytoplasm to exert its antitumor activity.
Novel function of PADI3 as a tumor suppressor gene in colon cancer
PADI3 belongs to the PAD family, and earlier studies have shown that it plays an important role in keratinocytes38,39.However, its role in tumorigenesis is not clear. In the present study, we found that it is highly expressed in adjacent tissues,but only weak PADI3 expression is detected in colon cancer tissues. In vitro, the overexpression of PADI3 in colon cancer cells reduced cell proliferation, induced G1 phase arrest and decreased colony formation. In vivo, PADI3-expressing HCT116 cells showed an obviously suppressed tumor formation ratio and growth, which indicates that PADI3 plays an antitumor role in colon cancer both in vitro and in vivo. This is the first report of the function of PADI3 in tumorigenesis as a tumor suppressor gene. The role of PADI3 in other types of cancer also needs to be investigated in future research.
PADI3 functions to induce G1 phase arrest by downregulating Sirt2 expression
Sirt2 regulates multiple cellular processes, including cell cycle progression, mitotic checkpoint control, autophagy and apoptosis17. Sirt2 inhibitors, such as AGK-2, AC-93253,AEM1, AEM2, AK1 and TM, can block colon cancer cell proliferation and migration in cervical cancer, prostate cancer, colon cancer, leukemia, glioma, and non-small-cell lung cancer17,23,27-31. An earlier report showed that the Sirt2 inhibitor AK-1 can suppress Snail expression and upregulate p21 expression, leading to G1 phase arrest in colon cancer17.Sirt2 inhibitors can also suppress cell migration and induce G1 phase arrest in hepatocellular carcinoma and colon cancer by suppressing AKT phosphorylation, inhibiting Snail expression and promoting p21 expression17,21. Our study found that the overexpression of PADI3 in HCT116 cells downregulated Snail expression, reduced AKT phosphorylation and upregulated p21 expression, which is in accordance with the findings of previous reports.Furthermore, we found that the overexpression of either Sirt2-iso1 or Sirt2-iso2 in PADI3-expressing HCT116 cells elevated AKT phosphorylation and Snail expression and suppressed p21 expression. We also found that the overexpression of Sirt2 in PADI3-expressing HCT116 cells rescued the inhibitory effect of PADI3 on the induction of G1 phase arrest. These results suggest that PADI3 can promote p21 expression and suppress AKT phosphorylation and Snail expression by inhibiting Sirt2 expression, which leads to cell cycle arrest and the inhibition of cell proliferation, ultimately suppressing colon cancer tumorigenesis. The low level of PADI3 expression in colonic tissue results in the loss of control of Sirt2 expression and contributes to tumorigenesis.
PADI3 regulates the expression of a set of genes in colon cancer
In this study, we found that the expression levels of Sirt2,U2AF1, SPON1, GADD45G, MSTN, RPS6KA1, PTPN22 and DIO2 were significantly altered by the overexpression of PADI3 at the transcriptional level. However, the molecular mechanism remains unknown. An increasing amount of evidence has shown that protein citrullination plays an important role in cancer pathogenesis40. PADI4, as another member of the PAD family, can catalyze arginine residues R2,R8, R17, and R26 in histone 3 tail citrullination and the R3 residue in histone 4 tail citrullination. This kind of histone posttranslational modification has the potential to antagonize histone methylation and coordinate with histone deacetylation to regulate gene transcription. We speculate that PADI3 regulates gene transcription via histone posttranslational modification. Further studies are needed to explore the effects and molecular mechanisms by which PADI3 affects these genes, which may help us further understand the function and underlying mechanisms of this enzyme in colon tumorigenesis.
C-domain is essential for the antitumor activity of PADI3 in the cytoplasm
Immunocytochemistry and overexpression experiments showed that PADI3 is mainly localized in the cytoplasm. A truncated mutation experiment indicated that the C-domain is essential for PADI3 to exert its antitumor activity in the cytoplasm. The C-domain, also called the PAD domain,exhibits similar antitumor activity to full-length PADI3 and can help PADI3 anchor in the cytoplasm. The identification of a shorter antitumor polypeptide and key antitumor sites is essential to explore its potential application value.
Conclusions
In vitro, PADI3 can inhibit cell proliferation, colony formation and lead to G1 phase arrest; in vivo, PADI3-expressing HCT116 cells had a lower tumor formation rate and produced smaller tumors than control cells. Molecular mechanism studies indicate that PADI3 suppresses Snail expression, AKT phosphorylation and promotes p21 expression by downregulating Sirt2 expression in the cytoplasm, and the C-domain is the key domain for PADI3 to play its antitumor activity.
Acknowledgments
This study was supported by the National Natural Science Foundation for the Youth of China (Grant No. 81802422 and 81702440), the Shandong Provincial Key R & D Program(Nos. 2019GSF108115, 2017GSF218102), Jinan Science and Technology Development Program (Nos. 201907116),Shandong Science and Technology Development Plan (Grant No. 2017GFS18195) and Shandong Traditional Chinese Medicine Science and Technology Development Programs(Grant No. 2017-173).
Conflict of interest statement
No potential conflicts of interest are disclosed.
 Cancer Biology & Medicine2019年4期
Cancer Biology & Medicine2019年4期
- Cancer Biology & Medicine的其它文章
- Interpretation of breast cancer screening guideline for Chinese women
- Breast cancer screening guideline for Chinese women
- Erratum to Simultaneous inhibition of PI3Kα and CDK4/6 synergistically suppresses KRAS-mutated non-small cell lung cancer
- The correlation and overlaps between PD-L1 expression and classical genomic aberrations in Chinese lung adenocarcinoma patients: a single center case series
- Nomogram based on albumin-bilirubin grade to predict outcome of the patients with hepatitis C virus-related hepatocellular carcinoma after microwave ablation
- Omics-based integrated analysis identified ATRX as a biomarker associated with glioma diagnosis and prognosis
