基于拟杆菌16S rRNA基因进行微生物溯源的研究进展
梁红霞,余志晟*,刘如铟,张洪勋,吴 钢
基于拟杆菌16S rRNA基因进行微生物溯源的研究进展
梁红霞1,余志晟1*,刘如铟1,张洪勋1,吴 钢2
(1.中国科学院大学资源与环境学院,北京 100049;2.中国科学院生态环境研究中心,北京 100085)
微生物溯源方法可利用粪便中的微生物区分来自人或动物的粪便污染.其中,拟杆菌以其丰度高、不能体外繁殖和宿主特异性强等优势被广泛应用于微生物溯源研究中.本文以拟杆菌16S rRNA基因为标记物,总结了拟杆菌及其标记物在环境中的衰减、拟杆菌引物的敏感性和特异性以及分子生物学技术在微生物溯源中的运用,可为粪便污染源解析提供一定的科学参考依据.
微生物溯源;拟杆菌;16S rRNA基因;粪便污染
近年来,随着我国畜牧业的快速发展,粪便的产生量日益增加.产生的粪便因管理不善或雨水冲刷等原因进入水中,容易引发一系列健康和环境问题.一方面,粪便中的病原菌会对公众健康造成一定威胁[1-2].研究表明粪便是水体病原菌的重要来源[3-4],水体病原菌可通过饮用水、贝类和水上娱乐活动等进行传播[5-6],进而增加传染病爆发的风险;另一方面,粪便引起的水体污染难以治理.未经处理的粪便N、P含量较高,易引起水体富营养化,使原有水体丧失功能[1,7].那么,如何区分人和动物的粪便污染对污染源的确定至关重要[8-10],而微生物溯源技术可实现快速有效区分.
微生物溯源(MST)[11-12]又称粪便污染溯源(FST),其原理是微生物与宿主肠道环境长期适应的过程中,逐渐形成了宿主专一性,并将这种专一性遗传给后代,使得这类微生物均具有某种特定的标记,通过这种特定的标记即可判断污染样品和可能污染物之间是否存在联系,从而实现粪便污染溯源[13-15].
在进行粪便污染溯源时,选择合适的粪便污染指示微生物至关重要.许多研究者推荐将拟杆菌作为微生物溯源的指示菌[16-17].拟杆菌是粪便中的优势菌.粪便干重的1/3为细菌,拟杆菌占粪便细菌总数的30%~40%[18-19].每克人粪便中的拟杆菌细胞数约为109~1011个,比肠杆菌或肠球菌高4~7个数量级[17,20-21];拟杆菌不能在体外环境中繁殖.拟杆菌进入水体后无法繁殖,故可通过其含量反映水体受污染的程度[16-17,22];拟杆菌具有宿主特异性.拟杆菌在与宿主长期适应的过程中,使其16S rRNA基因具有一定的宿主特异性.可利用拟杆菌16S rRNA基因判断污染样品和污染物之间的关系,进而确定污染源[23-24].
目前,大量研究表明以拟杆菌16S rRNA基因为标记物可成功区分哺乳动物、反刍动物和禽类等的粪便污染[24-26].本文在前人研究的基础上对相关内容进行归纳总结,以期为粪便污染的预防和微生物溯源技术在我国的发展提供一定帮助.
1 拟杆菌及其标记物的衰减
作为微生物溯源常用的指示菌,拟杆菌在宿主体外的存活时间备受关注,许多学者进行了相关研究.大量研究表明,拟杆菌在环境中的存活时间较短,有些粪便中的拟杆菌在环境水体中能存活数小时至数天[27-28],拟杆菌标记物则可持续数周[17,29-30],故可以很好的指示水体短期内发生的污染[16-17,30].
影响拟杆菌存活的因素较多,温度和捕食是主要因素,其次是光照和盐度.另外,水体营养程度、溶解氧含量和拟杆菌培养基质的状态等环境因素也会对拟杆菌及其标记物的衰减造成影响[31-33].
1.1 温度
研究发现温度越低,拟杆菌及其标记物的存在时间越长. Kreade[34]在河水中加入人的粪便模拟污水,发现拟杆菌标记物在4℃条件下可存在2周,14℃存在4~5d,而在24℃仅存在1~2d,30℃仅存在1d. Seurinck等[35]在厌氧条件下培养人粪便污水,发现拟杆菌标记物HF183在4和12℃的条件下可持续24d,而在28℃的条件下仅持续8d; Dick等[36]、Bae等[27]和Okabe等[37]在研究了不同宿主粪便来源的拟杆菌标记物后也得出了类似结论.
1.2 捕食
水体中的原生微生物或原生动物对拟杆菌标记物存在一定的捕食行为,且捕食行为越强,拟杆菌标记物的衰减速度越快.即水中活性因素的影响越大,拟杆菌标记物的存在时间越短. Bell等[38]将马粪和溪水混合后模拟粪便污水,分别研究了废水在过滤和不过滤两种情况下,拟杆菌标记物AllBac的衰减.研究发现将水样过滤后,拟杆菌标记物存在的时间更长. Dick等[36]研究发现捕食对人粪源拟杆菌标记物的影响比人工日光、沉积物和温度要大. Kreader[34]、Kobayashi等[39]及Ballesté等[31]的研究也一致认为捕食是影响拟杆菌标记物存在的主要因素.
1.3 光照
光照对拟杆菌标记物衰减速度的影响,研究结果并不一致,但这种不一致在其他微生物研究中也存在[40-41]. Bae等[27]研究了光照对4种拟杆菌及其标记物的影响.发现自然光并未明显地影响拟杆菌和其DNA的衰减(牛粪源拟杆菌标记物BacCow-UCD除外). Walters等[42]的发现与上述研究结果一致,该研究发现在光照和黑暗两种情况下,淡水中人粪源拟杆菌标记物HF134、HF183和反刍动物粪源标记物CF193的衰减行为并无明显差别.然而, Dick等[36]研究发现暴露在太阳光下会加速人粪源拟杆菌标记物qHF183和BacHum的衰减. Walters等[29]的研究也发现人粪源拟杆菌标记物BacHum的衰减速度在自然光下比黑暗中更快. Green等[43]的发现与Dick等[36]和Walters等[29]的结果一致.
1.4 盐度
盐度越高,拟杆菌标记物的衰减速度越慢. Green等[43]分别研究了人粪源拟杆菌标记物在海水和淡水中的衰减行为,结果表明拟杆菌标记物在海水中的持续时间比在淡水中长3d左右. Okabe等[37]研究了不同盐浓度的河水对通用拟杆菌、人粪源拟杆菌、牛粪源拟杆菌和猪粪源拟杆菌标记物存在时间的影响.这4种标记物的衰减行为一致说明盐度越高,拟杆菌标记物衰减速度越慢.值得说明的是此现象只出现在过滤后的水样,而在未过滤水样中则未出现,可能因为在未过滤水样中捕食是主要影响因素.
2 拟杆菌引物的敏感性和特异性
2.1 拟杆菌引物
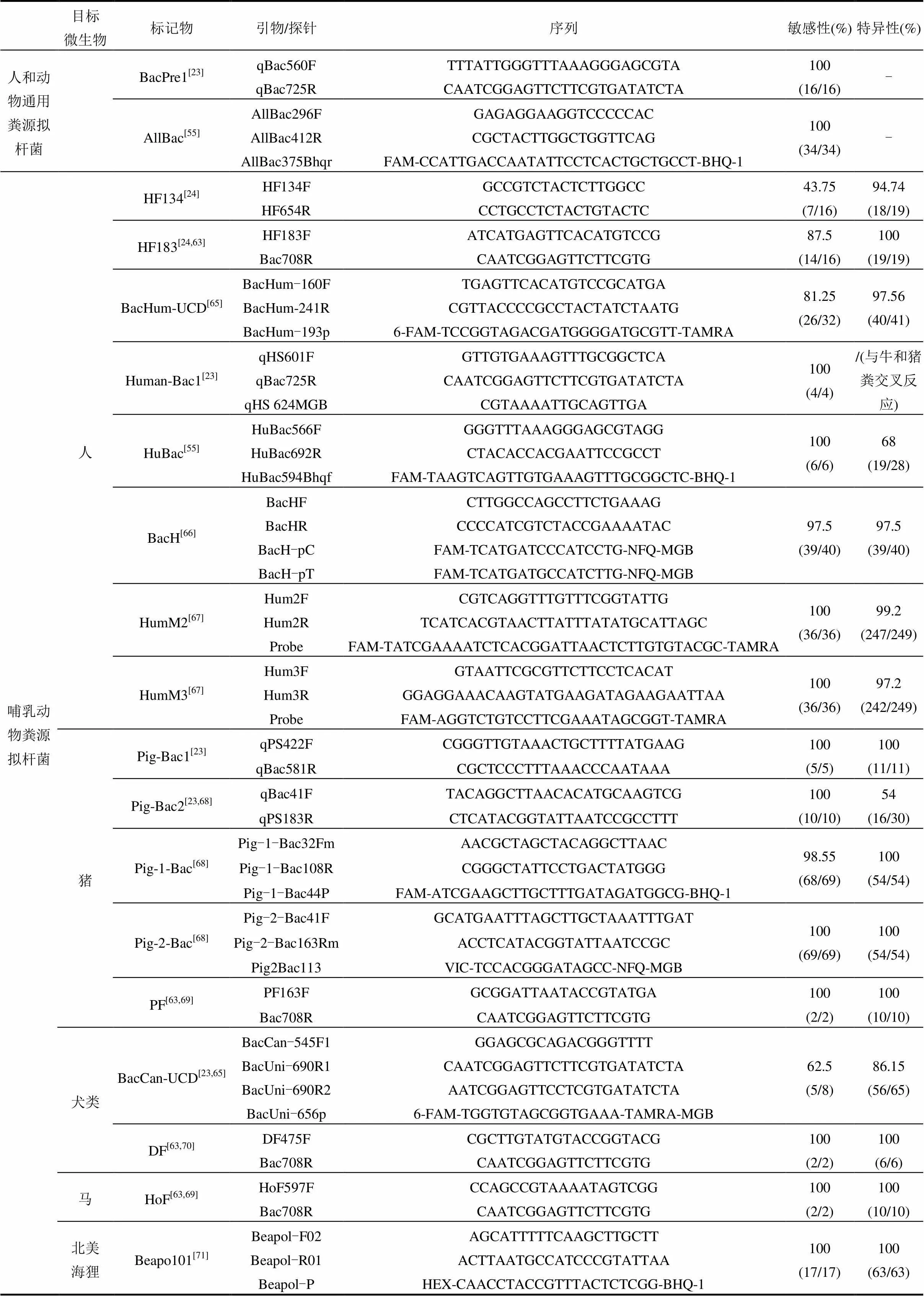
不同动物的饮食习惯、生活环境及肠道系统有所差异,拟杆菌在与宿主长期适应的过程中,使得其16S rRNA基因表现出一定的宿主特异性[24].利用拟杆菌16S rRNA基因构建系统发育树时,同种宿主粪源的拟杆菌群落表现出一定的相似性,不同宿主粪源的拟杆菌群落则出现分异,说明可根据拟杆菌标记物的存在指示不同宿主来源的粪便污染.通常使用拟杆菌引物判断标记物是否存在,拟杆菌引物分为拟杆菌通用引物和特异性引物.拟杆菌通用引物适用于多种动物,拟杆菌特异性引物则只针对某一种或某一类动物.
本节总结前人利用拟杆菌16S rRNA基因所设计的拟杆菌通用引物和特异性引物(表1),其中特异性引物针对的拟杆菌宿主包括:哺乳动物(人、猪、狗、马、北美海狸)、反刍动物(牛、麋鹿)及禽类(加拿大雁、鸡、鸭)等.目前,利用拟杆菌特异性标记物能够很好的区分人和反刍动物的粪便污染,但也存在一些问题,如关于禽类和水鸟粪便的溯源研究较少,主要有以下原因:(1)拟杆菌在有些禽类粪便中并不是优势菌[7].(2)禽类动物不仅包括城市中的禽类也包括野鸟,表现出高度多样的生境和饮食特征[44-45]. (3)不同的禽类会共同生活在同一片区域,无法获得单一性粪样[46].
2.2 引物的敏感性和特异性
拟杆菌标记物是否适合指示某种目标宿主的粪便污染,可用拟杆菌特异性引物的敏感性和特异性衡量.当引物的敏感性值=100%时,说明引物对应的拟杆菌标记物存在于所有目标宿主粪便样品中;特异性值=100%时,说明对应的拟杆菌标记物在所有非目标宿主粪便样品中均不存在.即拟杆菌引物敏感性和特异性值越高,对应的标记物越适合指示该宿主粪便的污染.
引物的敏感性()和特异性()可用下式表示[47]:
R
= TP/(TP + FN) (1)
S
= TN/(TN + FP) (2)
式中: TP(真阳性)为目标宿主显阳性的数量; FN(假阴性)为目标宿主显阴性的数量; TN(真阴性)为非目标宿主显阴性的数量; FP(假阳性)为非目标宿主显阳性的数量.
为了比较不同微生物溯源标记物的效果,世界范围内曾进行过3次大规模比选[48].(1) 2003年美国实验室间的比较研究[49-51];(2) 2006年欧洲实验室间的比较研究[52];(3) 2013年美国实验室间的比较研究[53-54].研究表明,拟杆菌通用引物Bac32F和Bac708R所对应的标记物存在于各种动物粪便中,可利用该引物对拟杆菌16S rRNA基因进行扩增后再进行后续实验[23,55-56];在多种人粪源拟杆菌标记物中, HF183表现出较强的敏感性和特异性[57-59];反刍动物标记物Rum-2-Bac和BacR在比选中表现出良好的特异性和敏感性,而且地理分布较广[54,60];但牛粪源拟杆菌标记物易与其他反刍动物或马发生交叉反应,说明牛粪源标记物也可能存在于其他反刍动物或马粪便中[54].
表1中的敏感性和特异性值由引物设计者实验得出,在一定程度上可为后续研究者在选择引物时提供参考.但不同拟杆菌标记物的地理分布有所差异,故不一定能重现这些数值. Reischer等[60]比较了人粪源标记物(BacH、BacHum)和反刍动物粪源标记物(BacCow、BacR和BoBac)在6个大洲的16个国家中的地理分布情况.5种标记物敏感性值分别是77%、87%、92%、90%和82%,说明牛粪源拟杆菌标记物地理分布最广;特异性值分别为53%、68%、57%、84%和59%,说明这5种标记物均存在交叉反应,即在非目标宿主中也存在. Shanks等[61]对2种反刍动物(CF128和CF193)和5种牛(Bac2、Bac3、BoBac、CowM2和CowM3)粪源拟杆菌标记物进行比较,结果表明反刍动物粪源标记物CF128和牛粪源标记物BoBac的敏感性最好且地理分布最广,但特异性较差,分别是76%和47.4%;牛粪源标记物Bac2、Bac3、CowM2和CowM3的特异性值均大于98.9%,但敏感性没有CF128和BoBac高. Tambalo等[62]对犬类粪便拟杆菌标记物CanBac-UCD进行了评估,只有31%的特异性,与设计者的86.15%相差较大.

表1 拟杆菌通用引物和特异性引物
续表1
目标微生物标记物引物/探针序列敏感性(%)特异性(%) 人和动物通用粪源拟杆菌BacPre1[23]qBac560FTTTATTGGGTTTAAAGGGAGCGTA100(16/16)- qBac725RCAATCGGAGTTCTTCGTGATATCTA AllBac[55]AllBac296FGAGAGGAAGGTCCCCCAC100(34/34)- AllBac412RCGCTACTTGGCTGGTTCAG AllBac375BhqrFAM-CCATTGACCAATATTCCTCACTGCTGCCT-BHQ-1 哺乳动物粪源拟杆菌人HF134[24]HF134FGCCGTCTACTCTTGGCC43.75(7/16)94.74(18/19) HF654RCCTGCCTCTACTGTACTC HF183[24,63]HF183FATCATGAGTTCACATGTCCG87.5(14/16)100(19/19) Bac708RCAATCGGAGTTCTTCGTG BacHum-UCD[65]BacHum-160FTGAGTTCACATGTCCGCATGA81.25(26/32)97.56(40/41) BacHum-241RCGTTACCCCGCCTACTATCTAATG BacHum-193p6-FAM-TCCGGTAGACGATGGGGATGCGTT-TAMRA Human-Bac1[23]qHS601FGTTGTGAAAGTTTGCGGCTCA100(4/4)/(与牛和猪粪交叉反应) qBac725RCAATCGGAGTTCTTCGTGATATCTA qHS 624MGBCGTAAAATTGCAGTTGA HuBac[55]HuBac566FGGGTTTAAAGGGAGCGTAGG100(6/6)68(19/28) HuBac692RCTACACCACGAATTCCGCCT HuBac594BhqfFAM-TAAGTCAGTTGTGAAAGTTTGCGGCTC-BHQ-1 BacH[66]BacHFCTTGGCCAGCCTTCTGAAAG97.5(39/40)97.5(39/40) BacHRCCCCATCGTCTACCGAAAATAC BacH-pCFAM-TCATGATCCCATCCTG-NFQ-MGB BacH-pTFAM-TCATGATGCCATCTTG-NFQ-MGB HumM2[67]Hum2FCGTCAGGTTTGTTTCGGTATTG100(36/36)99.2(247/249) Hum2RTCATCACGTAACTTATTTATATGCATTAGC ProbeFAM-TATCGAAAATCTCACGGATTAACTCTTGTGTACGC-TAMRA HumM3[67]Hum3FGTAATTCGCGTTCTTCCTCACAT100(36/36)97.2(242/249) Hum3RGGAGGAAACAAGTATGAAGATAGAAGAATTAA ProbeFAM-AGGTCTGTCCTTCGAAATAGCGGT-TAMRA 猪Pig-Bac1[23]qPS422FCGGGTTGTAAACTGCTTTTATGAAG100(5/5)100(11/11) qBac581RCGCTCCCTTTAAACCCAATAAA Pig-Bac2[23,68]qBac41FTACAGGCTTAACACATGCAAGTCG100(10/10)54(16/30) qPS183RCTCATACGGTATTAATCCGCCTTT Pig-1-Bac[68]Pig-1-Bac32FmAACGCTAGCTACAGGCTTAAC98.55(68/69)100(54/54) Pig-1-Bac108RCGGGCTATTCCTGACTATGGG Pig-1-Bac44PFAM-ATCGAAGCTTGCTTTGATAGATGGCG-BHQ-1 Pig-2-Bac[68]Pig-2-Bac41FGCATGAATTTAGCTTGCTAAATTTGAT100(69/69)100(54/54) Pig-2-Bac163RmACCTCATACGGTATTAATCCGC Pig2Bac113VIC-TCCACGGGATAGCC-NFQ-MGB PF[63,69]PF163FGCGGATTAATACCGTATGA100(2/2)100(10/10) Bac708RCAATCGGAGTTCTTCGTG 犬类BacCan-UCD[23,65]BacCan-545F1GGAGCGCAGACGGGTTTT62.5(5/8)86.15(56/65) BacUni-690R1CAATCGGAGTTCTTCGTGATATCTA BacUni-690R2AATCGGAGTTCCTCGTGATATCTA BacUni-656p6-FAM-TGGTGTAGCGGTGAAA-TAMRA-MGB DF[63,70]DF475FCGCTTGTATGTACCGGTACG100(2/2)100(6/6) Bac708RCAATCGGAGTTCTTCGTG 马HoF[63,69]HoF597FCCAGCCGTAAAATAGTCGG100(2/2)100(10/10) Bac708RCAATCGGAGTTCTTCGTG 北美海狸Beapo101[71]Beapol-F02AGCATTTTTCAAGCTTGCTT100(17/17)100(63/63) Beapol-R01ACTTAATGCCATCCCGTATTAA Beapol-PHEX-CAACCTACCGTTTACTCTCGG-BHQ-1
续表1

目标微生物标记物引物/探针序列敏感性(%)特异性(%) 反刍动物粪源拟杆菌反刍动物RUM[24,63]CF128FCCAACYTTCCCGWTACTC100(19/19)100(16/16) Bac708RCAATCGGAGTTCTTCGTG RUM[24,63]CF193FTATGAAAGCTCCGGCC100(19/19)100(16/16) Bac708RCAATCGGAGTTCTTCGTG Rum-2-Bac[56]BacB2-590FACAGCCCGCGATTGATACTGGTAA97(29/30)100(40/40) Bac708RmCAATCGGAGTTCTTCGTGAT BacB2-626PFAM-ATGAGGTGGATGGAATTCGTGGTGT-BHQ-1 BacR[72]BacR-FGCGTATCCAACCTTCCCG100(57/57)100(38/38) BacR-RCATCCCCATCCGTTACCG BacR-PFAM-CTTCCGAAAGGGAGATT-NFQ-MGB 牛Cow-Bac1[23]qCS406FGAAGGATGAAGGTTCTATGGATTGT100(7/7)100(9/9) qBac581RCGCTCCCTTTAAACCCAATAAA Cow-Bac2[23]qCS621FAACCACAGCCCGCGATT100(7/7)100(9/9) qBac725RCAATCGGAGTTCTTCGTGATATCTA Cow-Bac3[23]qBac41FTACAGGCTTAACACATGCAAGTCG100(7/7)100(9/9) qCS160RTCAACGGGCTATTCCTGAGTAAG BoBac[55]BoBac367FGAAG(G/A)CTGAACCAGCCAAGTA100(11/11)100(23/23) BoBac467RGCTTATTCATACGGTACATACAAG BoBac402PFAM-TGAAGGATGAAGGTTCTATGGATTGTAAACTT-BHQ-1 CowM2[73]CowM2FCGGCCAAATACTCCTGATCGT100(60/60)/ CowM2RGCTTGTTGCGTTCCTTGAGATAAT probeFAM-AGGCACCTATGTCCTTTACCTCATCAACTACAGACA-TAMRA CowM3[73]CowM3FCCTCTAATGGAAAATGGATGGTATCT100(60/60)/ CowM3RCCATACTTCGCCTGCTAATACCTT probeFAM-TTATGCATTGAGCATCGAGGCC-TAMRA BacCow-UCD[63,65]CF128FCCAACYTTCCCGWTACTC100(8/8)95.89(70/73) BacCow-305RGGACCGTGTCTCAGTTCCAGTG BacCow-257p6-FAM-TAGGGGTTCTGAGAGGAAGGTCCCCC-TAMRA 麋鹿EF[70]EF447FAATAACACCATCTACGTGTAGA100(2/2)80(8/10) EF990RGCCTGTCCAGTGCAATTTAA 禽类粪源拟杆菌鸡/鸭Chicken/Duck-Bac[26]qCD362F-HUAATATTGGTCAATGGGCGAGAG79.59(39/49)100(78/78) qcD464R-HUCACGTAGTGTCCGTTATTCCCTTA qBac394FAM-TCCTTCACGCTACTTGG-MGB 鸡Chicken-Bac[26]qC160F-HUAAGGGAGATTAATACCCGATGATG69.57(16/32)88.46(69/78) qBac265R-HUCCGTTACCCCGCCTACTAC 鸭Duck-Bac[26]qBac366F-HUTTGGTCAATGGGCGGAAG84.62(22/26)94.87(74/78) qDuck474R-HUGCACATTCCCACACGTGAGA qBac394FAM-TCCTTCACGCTACTTGG-MGB 加拿大雁CGOF1-Bac[74]CG1FGTAGGCCGTGTTTTAAGTCAGC57.43(58/101)100(291/291) CG1RAGTTCCGCCTGCCTTGTCTA ProbeFAM-CCGTGCCGTTATACTGAGACACTTGAG-BHQ-1 CGOF2-Bac[74]CG2FACTCAGGGATAGCCTTTCGA50.50(51/101)100(291/291) CG2RACCGATGAATCTTTCTTTGTCTCC ProbeFAM-AATACCTGATGCCTTTGTTTCCCTGCA-BHQ-1
注:/为未命名或数据未提供;-为数据不存在.
3 分子生物学技术在微生物溯源中的运用
在微生物溯源研究中,PCR技术常与其他技术结合使用以实现对污染物的定性分析,实时荧光定量PCR技术则常用于污染物的定量分析;基因芯片最大的特点是通量高,可同时检测成千上万个样品,成功识别污染源的同时还可对样品中微生物多样性进行分析.以上方法各有优缺点,在实际研究工作中,可根据实验目的选择合适的方法,还可同时使用多种方法以增加实验的可信度[75-77].
3.1 PCR及其联用技术
3.1.1 PCR主要操作如下 使用拟杆菌引物进行PCR扩增,观察目标条带是否出现.使用拟杆菌通用引物时,出现目标条带则说明样品存在粪便污染;使用拟杆菌特异性引物时,出现目标条带则说明样品中存在特异性粪便污染.目前该方法已成功地判断出水样被何种动物的粪便污染[24,63].但PCR技术也存在一定的缺陷,如易产生非特异性扩增,只能对污染物定性而不能定量等.
3.1.2 末端标记限制性酶切长度多态性(T- RFLP) 同一目的基因由于碱基的插入、缺失、重排或点突变,故在不同的微生物间存在长度多态性[78].用带有荧光标记的引物扩增DNA样品,然后用限制性核酸内切酶酶切扩增产物,由于不同微生物的同一基因的核苷酸序列存在差异,所以同一个基因的DNA片段经相同酶切后可能得到长度不同的限制性内切片段,在T-RFLP检测中表现为不同的荧光信号[79]. Bernhard等[24,63]根据T-RFLP图谱区别出人和牛粪便中的拟杆菌16S rRNA基因特异性片段,并据此设计了拟杆菌通用引物和特异性引物. Dick等[69]在设计拟杆菌引物时,也用到了T-RFLP技术.
3.1.3 变性梯度凝胶电泳(DGGE) 利用菌株的标记基因获得样品的特异性指纹图谱,以指纹图谱的差异表征菌株的差异,进而分析被污染样品与污染物之间的关系.目前, DGGE技术在以大肠杆菌为指示菌进行微生物溯源方面研究较多[80-82],而以拟杆菌为指示菌进行微生物溯源方面的相关研究国内外鲜有报道.张曦等[83]利用拟杆菌特异性16S rRNA基因和大肠杆菌特异性基因phoE这两种标记基因,经DGGE技术对塘坝型饮用水污染进行溯源,研究结果显示拟杆菌DGGE图谱比大肠杆菌图谱的条带更丰富,且样品之间的显著性更高,表明可利用拟杆菌的DGGE图谱表征污染水体之间的关系.
3.2 实时荧光定量PCR技术(qPCR)
qPCR技术相比常规PCR技术有以下优点:(1) qPCR技术可对污染物进行定量分析,即回答污染物类型和污染物的量,而常规PCR技术只能对污染物进行定性分析,即回答污染物类型;(2) qPCR技术的灵敏性和特异性更好.基于 qPCR技术的优势,其在微生物溯源中有更广泛的运用,主要有以下3点:
3.2.1 区分不同类型的粪便污染并对污染物定量 Seurinck等[35]首次利用人粪源拟杆菌HF183标记物对水中的人粪源污染进行研究并取得很好的定量效果. Okabe等[23]选取了1种人粪源、3种牛粪源和2种猪粪源的拟杆菌标记物进行溯源实验,成功地判断出河水的粪便污染来源并测定出污染量. Jeong等[84]使用人粪源和牛粪源拟杆菌标记物进行TaqMan qPCR实验,结果表明利用qPCR技术能可靠地识别和量化粪便污染,为流域水质管理和改进方面提供有效的信息.
3.2.2 验证标记物的敏感性和特异性 Raith等[85]检验了5种拟杆菌标记物是否适用于加利福尼亚地区反刍动物的粪便污染.研究表明将牛粪源拟杆菌标记物CowM2和反刍动物粪源拟杆菌标记物BacR或Rum2Bac结合使用,最适合该地区反刍动物的粪便污染检验. Mieszkin等[68]使用两种猪粪源拟杆菌标记物评估了养猪场下游水域污染. qPCR实验结果表明这两种标记物具有良好的敏感性和特异性,可用于检验水环境中猪粪便的污染. Lee等[86]使用通用、人粪源和牛粪源拟杆菌标记物分别进行TaqMan qPCR实验,验证了这3种标记物具有良好的特异性和敏感性.
3.2.3 发现粪便中的优势菌 Matsuki等[19,87]利用多种特异性引物进行qPCR实验,表明脆弱拟杆菌是人粪便中的优势菌.
3.3 基因芯片技术
基因芯片又称DNA芯片或DNA微阵列,原理是采用光导原位合成或显微印刷等方法将大量特定序列的探针分子密集、有序地固定于经过相应处理的硅片、玻片、硝酸纤维素膜等载体上,然后加入标记的待测样品,进行多元杂交,通过杂交信号的强弱及分布,来分析目的分子的有无、数量及序列,从而获得受检样品的遗传信息[88].
基因芯片技术在微生物溯源中表现出一定的可靠性[89-93],主要应用在以下两方面:
3.3.1 评估水体污染状况 Dubinsky等[94]采集了42个包括人、鸟、牛、马、麋鹿和鳍足类动物的粪便排泄物,使用59316种不同的细菌16S rRNA基因探针进行检测.其中梭状芽胞杆菌门和拟杆菌门中的多种科能把人、食草动物和鳍足亚目类动物3者的污染区分开,为加利福尼亚沿海水域提供污染源信息. Inoue等[95]调查加德满都谷地的浅井地下水污染状况时,以941个病原菌为对象进行基因芯片实验,证明该地区的浅井地下水普遍受到粪便污染.
3.3.2 分析粪便样品中微生物多样性 Li等[96]在前人研究基础上,利用鸡、牛、家禽和猪粪便中微生物的DNA或RNA,将基因芯片、qPCR技术及二代测序技术相结合,检测到不同粪便中含有相同病原菌且病原菌主要有两类:和. Wang等[90]使用CY-5荧光基团标记人粪便中的肠道细菌的16S rRNA基因,荧光杂交结果说明粪便中主要的肠道细菌是普通拟杆菌、梭形杆菌属、多型拟杆菌、瘤胃球菌属、消化链球菌和真杆菌,与前人的研究结果一致[97-98].
基因芯片在微生物溯源研究中有许多优点.如:(1)通量高,可同时检测成千上百个样品,全面分析样品中的致病微生物;(2)检测速度快;(3)在样品量很少的情况下仍能保持高敏感性.当然,它也存在一定的缺陷.如:(1)以16S rRNA基因为待测基因时,16S rRNA基因数据库不足,导致探针设计存在缺陷;(2)成本和操作复杂度高.总体来说,基因芯片技术是一种快速有效判定污染物来源的技术,具有广阔的应用空间.
4 结语
近年来,利用拟杆菌16S rRNA基因对粪便污染进行溯源已经取得了很大的进展,但仍面临一些挑战,现做如下总结和展望:
4.1 拟杆菌标记物可实现对人和反刍动物粪便污染的准确溯源,但拟杆菌在一些禽类和鸟类粪便中并不是优势菌,故利用拟杆菌对禽类粪便污染溯源时,效果并不理想.需进一步发现其他标记物以实现鸟类和禽类粪便污染的准确溯源.
4.2 拟杆菌是严格厌氧菌,在水体中的存在量随时间逐渐减少,适用于指示近期发生的污染,但无法对污染时间较长的水体进行评估,故在实际研究中可将拟杆菌标记物与其他标记物,如线粒体DNA结合使用.
4.3 需尽快建立拟杆菌标记物含量与水体污染程度之间的对应关系,为政府问责及司法鉴定提供依据.
4.4 各国实验室应加强合作,积极开展拟杆菌的地理分布研究,在世界范围内确定敏感性和特异性效果最好的拟杆菌标记物.
[1] Harwood V J, Staley C, Badgley B D, et al. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes [J]. Fems Microbiology Reviews, 2013,38(1):1-40.
[2] Samadpour M, Stewart J, Steingart K, et al. Laboratory investigation of anO157: H7outbreak associated with swimming in Battle Ground Lake, Vancouver, Washington [J]. Journal of Environmental Health, 2002,64(10):16-20.
[3] Hamilton M J, Yan T, Sadowsky M J. Development of goose- and duck-specific DNA markers to determine sources ofin waterways [J]. Applied and Environmental Microbiology, 2006,72(6):4012-4019.
[4] Santo Domingo J W, Bambic D G, Edge T A, et al. Quo vadis source tracking? Towards a strategic framework for environmental monitoring of fecal pollution [J]. Water Research, 2007,41(16):3539- 3552.
[5] Alexander L M, Heaven A, Tennant A, et al. Symptomatology of children in contact with sea water contaminated with sewage [J]. Journal of Epidemiology and Community Health, 1992,+46(4):340- 344.
[6] US Department of Health and Human Services, Center for Disease Control and Prevention. Outbreak ofparahaemolyticus infections associated with eating raw oysters-Pacific Northwest, 1997 [J]. Morbidity and Mortality Weekly Report, 1998,47(22):457-462.
[7] Lu J R, Domingo J S, Shanks O C. Identification of chicken-specific fecal microbial sequences using a metagenomic approach [J]. Water Research, 2007,41(16):3561-3574.
[8] Ahmed W, Sritharan T, Palmer A, et al. Evaluation of bovine feces- associated microbial source tracking markers and their correlations with fecal indicators and zoonotic pathogens in a Brisbane, Australia, reservoir [J]. Applied and Environmental Microbiology, 2013,79(8): 2682-2691.
[9] Lu J R, Ryu H, Vogel J, et al. Molecular detection ofspp. and fecal indicator bacteria during the northern migration of sandhill cranes () at the central platte river [J]. Applied and Environmental Microbiology, 2013,79(12):3762-3769.
[10] Mclellan S L, Eren A M. Discovering new indicators of fecal pollution [J]. Trends in Microbiology, 2014,22(12):697-706.
[11] Wiggins B A. Discriminant analysis of antibiotic resistance patterns in fecal, a method to differentiate human and animal sources of fecal pollution in natural waters [J]. Applied and Environmental Microbiology, 1996,62(11):3997-4002.
[12] Hagedorn C, Robinson S L, Filtz J R, et al. Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal[J]. Applied and Environmental Microbiology, 1999,65(12):5522-5531.
[13] Simpson J M, Santo Domingo J W, Reasoner D J. Microbial source tracking: state of the science [J]. Environmental Science and Technology, 2002,36(24):5279-5288.
[14] Scott T M, Rose J B, Jenkins T M, et al. Microbial source tracking: current methodology and future directions [J]. Applied and Environmental Microbiology, 2002,68(12):5796-5803.
[15] 冯广达,邓名荣,郭 俊,等.广东某农村塘坝饮用水污染的微生物溯源 [J]. 中国环境科学, 2011,31(1):96-104.
[16] Allsop K, Stickler D J. An assessment offragilis group organisms as indicators of human faecal pollution [J]. Journal of Applied Microbiology, 1985,58(1):95-99.
[17] Fiksdal L, Maki J S, Lacroix S J, et al. Survival and detection ofspp. prospective indicator bacteria [J]. Applied and Environmental Microbiology, 1985,49(1):148-150.
[18] Hayashi H, Sakamoto M, Benno Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods [J]. Microbiology and Immunology, 2002,46(8):535-548.
[19] Matsuki T, Watanabe K, Fujimoto J, et al. Use of 16s rRNA gene- targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces [J]. Applied and Environmental Microbiology, 2004,70(12):7220-7228.
[20] Hold G L, Pryde S E, Russell V J, et al. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis [J]. Fems Microbiology Ecology, 2002,39(1):33-39.
[21] Leser T D, Amenuvor J Z, Jensen T K, et al. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited [J]. Applied and Environmental Microbiology, 2002,68(2): 673-690.
[22] Avelar K E, Moraes S R, Pinto L J, et al. Influence of stress conditions onfragilis survival and protein profiles [J]. Zentralblatt Für Bakteriologie, 1998,287(4):399-409.
[23] Okabe S, Okayama N, Savichtcheva O, et al. Quantification of host-specific, 16S rRNA genetic markers for assessment of fecal pollution in freshwater [J]. Applied Microbiology and Biotechnology, 2007,74(4):890-901.
[24] Bernhard A E, Field K G. A PCR assay to discriminate human and ruminant feces on the basis of host differences in-genes encoding 16S rRNA [J]. Applied and Environmental Microbiology, 2000,66(10):4571-4574.
[25] 张 杨,吴仁人,张一敏,等.珠三角河网地区粪便污染源解析 [J]. 中国环境科学, 2017,37(9):3446-3454.
[26] Kobayashi A, Sano D, Hatori J, et al. Chicken- and duck-associated, genetic markers for detecting fecal contamination in environmental water [J]. Applied Microbiology and Biotechnology, 2013,97(16):7427-7437.
[27] Bae S, Wuertz S. Rapid decay of host-specific fecalcells in seawater as measured by quantitative PCR with propidium monoazide [J]. Water Research, 2009,43(19):4850-4859.
[28] Liang Z B, He Z L, Zhou X X, et al. High diversity and differential persistence of fecalpopulation spiked into freshwater microcosm [J]. Water Research, 2012,46(1):247-257.
[29] Walters S P, Yamahara K M, Boehm A B. Persistence of nucleic acid markers of health-relevant organisms in seawater microcosms: implications for their use in assessing risk in recreational waters [J]. Water Research, 2009,43(19):4929-4939..
[30] Tambalo D D, Fremaux B, Boa T, et al. Persistence of host-associatedgene markers and their quantitative detection in an urban and agricultural mixed prairie watershed [J]. Water Research, 2012,46(9):2891-2904.
[31] Ballesté E, Blanch A R. Persistence ofspecies populations in a river as measured by molecular and culture techniques [J]. Journal of Applied Microbiology, 2010,76(22):7608-7616.
[32] Rocha E R, Herren C D, Smalley D J, et al. The complex oxidative stress response offragilis: the role of OxyR in control of gene expression [J]. Anaerobe, 2003,9(4):165-173.
[33] Brooks Y, Aslan A, Tamrakar S, et al. Analysis of the persistence of enteric markers in sewage polluted water on a solid matrix and in liquid suspension [J]. Water Research, 2015,76:201-212.
[34] Kreader C A. Persistence of PCR-detectable Bacteroides distasonis from human feces in river water [J]. Applied and Environmental Microbiology.1998,64(10):4103-4105.
[35] Seurinck S, Defoirdt T, Verstraete W, et al. Detection and quantification of the human-specific HF183, 16S rRNA genetic marker with real-time PCR for assessment of human faecal pollution in freshwater [J]. Environmental Microbiology, 2005,7(2): 249-259.
[36] Dick L K, Stelzer E A, Bertke E E, et al. Relative decay ofmicrobial source tracking markers and cultivatedin freshwater microcosms [J]. Applied and Environmental Microbiology, 2010,76(10):3255-3262.
[37] Okabe S, Shimazu Y. Persistence of host-specific16S rRNA genetic markers in environmental waters: effects of temperature and salinity [J]. Applied Microbiology and Biotechnology, 2007,76(4):935-944.
[38] Bell A, Layton A C, Mckay L, et al. Factors influencing the persistence of fecalin stream water [J]. Journal of Environmental Quality, 2009,38(3):1224-1232.
[39] Kobayashi A, Sano D, Okabe S. Effects of temperature and predator on the persistence of host-specificgenetic markers in water [J]. Water Science and Technology A Journal of the International Association on Water Pollution Research, 2013,67(4): 838-845.
[40] Barcina I, Lebaron P, VivesRego J. Survival of allochthonous bacteria in aquatic systems: a biological approach [J]. Fems Microbiology Ecology, 1997,23(1):1–9.
[41] Rozen Y, Belkin S. Survival of enteric bacteria in seawater [J]. Fems Microbiology Reviews, 2001,25(5):513-529.
[42] Walters S P, Field K G. Survival and persistence of human and ruminant-specific faecalin freshwater microcosms [J]. Environmental Microbiology, 2009,11(6):1410–1421.
[43] Green H C, Shanks O C, Sivaganesan M, et al. Differential decay of human faecalin marine and freshwater [J]. Environmental Microbiology, 2011,13(12):3235-3249.
[44] Zhu X Y, Zhong T Y, Pandya Y, et al. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens [J]. Applied and Environmental Microbiology, 2002,68(1):124-137.
[45] Xenoulis P G, Gray P L, Brightsmith D, et al. Molecular characterization of the cloacal microbiota of wild and captive parrots [J]. Veterinary Microbiology, 2010,146(3/4):320-325.
[46] Ohad S, Ben-Dor S, Prilusky J, et al. The development of a novel qPCR assay-set for identifying fecal contamination originating from domestic fowls and waterfowl in Israel [J]. Frontiers in Microbiology, 2016,7(e893378):145.
[47] Ballesté E, Bonjoch X, Belanche L A, et al. Molecular indicators used in the development of predictive models for microbial source tracking [J]. Applied and Environmental Microbiology, 2010,76(6):1789-1795.
[48] 魏源送,郑嘉熹,王光宇,等.地表水微生物溯源技术的开发和应用进展 [J]. 水资源保护, 2016,32(1):1-11.
[49] Field K G, Chern E C, Dick L K, et al. A comparative study of culture-independent, library-independent genotypic methods of fecal source trackin [J]. Journal of Water and Health, 2003,1(4):181-194.
[50] Harwood V J, Wiggins B, Hagedorn C, et al. Phenotypic library-based microbial source tracking methods: efficacy in the California collaborative study [J]. Journal of Water and Health, 2003,1(4):153- 166.
[51] Myoda S P, Carson C A, Fuhrmann J J, et al. Comparison of genotypic-based microbial source tracking methods requiring a host origin database [J]. Journal of Water and Health, 2003,1(4):167-180.
[52] Blanch A R, Belanche-Munoz L, Bonjoch X, et al. Integrated analysis of established and novel microbial and chemical methods for microbial source tracking [J]. Applied and Environmental Microbiology, 2006, 72(9):5915-5926.
[53] Stewart J R, Boehm A B, Dubinsky E A, et al. Recommendations following a multi-laboratory comparison of microbial source tracking methods [J]. Water Research, 2013,47(18):6829-6838.
[54] Boehm A B, Van De Werfhorst L C, Griffith J F, et al. Performance of forty-one microbial source tracking methods: A twenty-seven lab evaluation study [J]. Water Research, 2013,47(18):6812-6828.
[55] Layton A, Mckay L, Williams D, et al. Development of16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water [J]. Applied and Environmental Microbiology, 2006,72(6):4214-4224.
[56] Mieszkin S, Yala J F, Joubrel R, et al. Phylogenetic analysis of16S rRNA gene sequences from human and animal effluents and assessment of ruminant faecal pollution by real-time PCR [J]. Journal of Applied Microbiology, 2010,108(3):974–984.
[57] Ahmed W, Goonetilleke A, Powell D, et al. Comparison of molecular markers to detect fresh sewage in environmental waters [J]. Water Research, 2009,43(19):4908-4917.
[58] Ahmed W, Masters N, Toze S. Consistency in the host specificity and host sensitivity of the, HF183marker for sewage pollution tracking [J]. Letters in Applied Microbiology, 2012,55(4):283–289.
[59] Layton B A, Cao Y, Ebentier D L, et al. Performance of human fecal anaerobe-associated PCR-based assays in a multi-laboratory method evaluation study [J]. Water Research, 2013,47(18):6897-6908.
[60] Reischer G H, Ebdon J E, Bauer J M, et al. Performance characteristics of qPCR assays targeting human- and ruminant- associatedfor microbial source tracking across sixteen countries on six continents [J]. Environmental Science and Technology, 2013,47(15):8548-8556.
[61] Shanks O C, White K, Kelty C A, et al. Performance assessment PCR-based assays targetinggenetic markers of bovine fecal pollution [J]. Applied and Environmental Microbiology, 2010,76(5):1359-1366.
[62] Tambalo D D, Boa T, Liljebjelke K, et al. Evaluation of two quantitative PCR assays usingand mitochondrial DNA markers for tracking dog fecal contamination in waterbodies [J]. Journal of Microbiological Methods, 2012,91(3):459-467.
[63] Bernhard A E, Field K G. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16s ribosomal DNA genetic markers from fecal anaerobes [J]. Applied and Environmental Microbiology, 2000,66(4):1587-1594.
[64] Manz W, Amann R, Ludwig W, et al. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylumin the natural environment [J]. Microbiology, 1996,142(Pt5):1097-1106.
[65] Kildare B J, Leutenegger C M, Mcswain B S, et al. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal: a Bayesian approach [J]. Water Research, 2007,41(16):3701-3715.
[66] Reischer G H, Kasper D C, Steinborn R, et al. A quantitative real-time PCR assay for the highly sensitive and specific detection of human faecal influence in spring water from a large alpine catchment area [J]. Letters in Applied Microbiology, 2007,44(4):351-356.
[67] Shanks O C, Kelty C A, Sivaganesan M, et al. Quantitative PCR for genetic markers of human fecal pollution [J]. Applied and Environmental Microbiology, 2009,75(17):5507-5513.
[68] Mieszkin S, Furet J P, Corthier G, et al. Estimation of pig fecal contamination in a river catchment by real-time PCR using two pig-specific16S rRNA genetic markers [J]. Applied and Environmental Microbiology, 2009,75(10):3045-3054.
[69] Dick L K, Bernhard A E, Brodeur T J, et al. Host distributions of uncultivated fecalbacteria reveal genetic markers for fecal source identification [J]. Applied and Environmental Microbiology, 2005,71(6):3184-3191.
[70] Dick L K, Simonich M T, Field K G. Microplate subtractive hybridization to enrich forgenetic markers for fecal source identification [J]. Applied and Environmental Microbiology, 2005,71(6):3179-3183.
[71] Marti R, Zhang Y, Tien Y-C, et al. Assessment of a newmarker targeting North American beaver () fecal pollution by real-time PCR [J]. Journal of Microbiological Methods, 2013,95(2):201-206.
[72] Reischer G H, Kasper D C, Steinborn R, et al. Quantitative PCR method for sensitive detection of ruminant fecal pollution in freshwater and evaluation of this method in alpine karstic regions [J]. Applied and Environmental Microbiology, 2006,72(8):5610-5614.
[73] Shanks O C, Atikovic E, Blackwood A D, et al. Quantitative PCR for detection and enumeration of genetic markers of bovine fecal pollution [J]. Applied and Environmental Microbiology, 2008,74(3):745-752.
[74] Fremaux B, Boa T, Yost C K. Quantitative real-time PCR assays for sensitive detection of Canada goose-specific fecal pollution in water sources [J]. Applied and Environmental Microbiology, 2010,76(14): 4886-4889.
[75] Roslev P, Bukh A S. State of the art molecular markers for fecal pollution source tracking in water [J]. Applied Microbiology and Biotechnology, 2011,89(5):1341-1355.
[76] Lee D Y, Lauder H, Cruwys H, et al. Development and application of an oligonucleotide microarray and real-time quantitative PCR for detection of wastewater bacterial pathogens [J]. Science of the Total Environment, 2008,398(1-3):203-211.
[77] Lee D Y, Shannon K, Beaudette L A. Detection of bacterial pathogens in municipal wastewater using an oligonucleotide microarray and real-time quantitative PCR [J]. Journal of Microbiological Methods, 2006,65(3):453-467.
[78] Suzuki M, Rappe M S, Giovannoni S J. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity [J]. Applied and Environmental Microbiology, 1998,64(11):4522-4529.
[79] Liu W T, Marsh T L, Cheng H, et al. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA [J]. Applied and Environmental Microbiology, 1997,63(11):4516-4522.
[80] Farnleitner A H, Kreuzinger N, Kavka G G, et al. Simultaneous detection and differentiation ofpopulations from environmental freshwaters by means of sequence variations in a fragment of the beta-D-glucuronidase gene [J]. Applied and Environmental Microbiology, 2000,66(4):1340-1346.
[81] Ram J L, Ritchie R P, Fang J, et al. Sequence-based source tracking ofbased on genetic diversity of beta-glucuronidase [J]. Journal of Environmental Quality, 2004,33(3):1024-1032.
[82] Esseili M A, Kassem I I, Sigler V. Optimization of DGGE community fingerprinting for characterizing, communities associated with fecal pollution [J]. Water Research, 2008,42(17): 4467-4476.
[83] 张 曦,朱昌雄,冯广达,等.基于拟杆菌特异性16S rRNA基因的塘坝型饮用水污染溯源研究 [J]. 农业环境科学学报, 2011,30(9): 1880-1887.
[84] Jeong J Y, Park H D, Lee K H, et al. Quantitative analysis of human- and cow-specific 16s rRNA gene markers for assessment of fecal pollution in river waters by real-time PCR [J]. Journal of Microbiology and Biotechnology, 2010,20(2):245-253.
[85] Raith M R, Kelty C A, Griffith J F, et al. Comparison of PCR and quantitative real-time PCR methods for the characterization of ruminant and cattle fecal pollution sources [J]. Water Research, 2013, 47(18):6921-6928.
[86] Lee D Y, Weir S C, Lee H, et al. Quantitative identification of fecal water pollution sources by TaqMan real-time PCR assays using16S rRNA genetic markers [J]. Applied Microbiology and Biotechnology, 2010,88(6):1373-1383.
[87] Matsuki T, Watanabe K, Fujimoto J, et al. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces [J]. Applied and Environmental Microbiology, 2002,68(11):5445-5451.
[88] Bao N, Kassegne S K. DNA Microarrays [J]. Nanoscience Nanobiotechnology and Nanobiology, 2008,109(1):433-453.
[89] Indest K J, Betts K, Furey J S. Application of oligonucleotide microarrays for bacterial source tracking of environmentalsp. Isolates [J]. International Journal of Environmental Research and Public Health, 2005,2(1):175-185.
[90] Wang R F, Beggs M L, Robertson L H, et al. Design and evaluation of oligonucleotide-microarray method for the detection of human intestinal bacteria in fecal samples [J]. Fems Microbiology Letters, 2002,213(2):175-182.
[91] Straub T M, Daly D S, Wunshel S, et al. Genotypingwith an70single-nucleotide polymorphism microarray [J]. Applied and Environmental Microbiology, 2002,68(4):1817-1826.
[92] Maynard C, Berthiaume F, Lemarchand K, et al. Waterborne pathogen detection by use of oligonucleotide-based microarrays [J]. Applied and Environmental Microbiology, 2005,71(12):8548-8557.
[93] Li X, Harwood V J, Nayak B, et al. Ultrafiltration and microarray for detection of microbial source tracking marker and pathogen genes in riverine and marine systems [J]. Applied and Environmental Microbiology, 2016,82(5):1625-1635.
[94] Dubinsky E A, Esmaili L, Hulls J R, et al. Application of phylogenetic microarray analysis to discriminate sources of fecal pollution [J]. Environmental Science and Technology, 2012,46(8):4340-4347.
[95] Inoue D, Hinoura T, Suzuki N, et al. High-throughput DNA microarray detection of pathogenic bacteria in shallow well groundwater in the Kathmandu Valley, Nepal [J]. Current Microbiology, 2015,70(1):43-50.
[96] Li X, Harwood V J, Nayak B, et al. A novel microbial source tracking microarray for pathogen detection and fecal source identification in environmental systems [J]. Environmental Science and Technology, 2015,49(12):7319-7329.
[97] Moore W E C, Holdeman L V. Human fecal flora: the normal flora of 20Japanese-Hawaiians [J]. Applied Microbiology, 1974,27(5):961- 979.
[98] Cerniglia C E, Kotarski S. Evaluation of veterinary drug residues in food for their potential to affect human intestinal microflora [J]. Regulatory Toxicology and Pharmacology, 1999,29(3):238-261.
致谢:感谢中国科学院大学常冬冬博士对本论文的审阅和修订.
Research progress of microbial source tracking based on16S rRNA gene.
LIANG Hong-xia1, YU Zhi-sheng1*, LIU Ru-yin1, ZHANG Hong-xun1, WU Gang2
(1.College of Resources and Environment, University of Chinese Academy of Sciences, Beijing 100049, China;2.Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences, Beijing 100085, China)., 2018,38(11):4236~4245
Microbial source tracking (MST) is a method for distinguishing fecal contamination from different animals by identifying specific fecal microbes.is widely used in MST because of its high abundance in feces, non-reproduction, and host specificity. Given thats 16S rRNA gene is a common biomarker for MST, this paper reviewed the decay ofand its biomarkers, the sensitivity and specificity of the 16S rRNA gene primers for, and the application of molecular techniques in MST. It will provide the appropriate scientific reference for the source apportionment of feces.
microbial source tracking;;16S rRNA gene;fecal pollution
X172
A
1000-6923(2018)11-4236-10
梁红霞(1990-),女,河南鹿邑人,硕士研究生,主要从事环境微生物方向的研究.
2018-04-08
国家重点研发计划(2016YFC0503601);中国科学院战略性先导科技专项(B类)(XDB15010200)
*责任作者, 教授, yuzs@ucas.ac.cn

