鹅豁眼性状H基因座候选基因FREM1的验证分析
于金成,于宁,赵辉,李喆
(辽宁省农业科学院草牧业研究所,沈阳 110161)
鹅豁眼性状H基因座候选基因FREM1的验证分析
于金成,于宁,赵辉,李喆
(辽宁省农业科学院草牧业研究所,沈阳 110161)
【目的】鹅豁眼性状呈隐性伴性遗传,其遗传基础有待揭示。基因FRAS-related extracellular matrix 1(FREM1)编码区的一些隐性突变导致了人类及模型小鼠上眼睑部分或完全缺失。本试验以鹅豁眼性状资源群为主要材料,通过对鹅基因FREM1的克隆、表达及基因多态性分析,验证FREM1是影响鹅上眼睑性状候选基因的假设,为深入研究眼睑性状的分子遗传机制奠定基础。【方法】采集鹅豁眼性状F2资源群中成年纯种豁眼鹅母鹅(3只)、四川白鹅(6只,雌雄各半)、♂豁眼×♀四川白鹅F1代公鹅(3只)的眼睑和肾组织,提取其总RNA,以鹅FREM1(XM_013193557)全长转录序列为参考设计引物,利用反转录RT-PCR克隆鹅FREM1基因,对其进行生物信息学分析,进而,采用实时荧光定量PCR法研究鹅眼睑和肾组织FREM1基因的表达特性。采集成年纯种豁眼鹅公鹅、四川白鹅公鹅和♂豁眼×♀四川白鹅正反交F1代公鹅各30只的血液,提取全血DNA,以鹅FREM1(Anser cygnoides domesticus breed Zhedong scaffold203_32,NCBI)序列设计引物,利用直接测序法检测FREM1基因变异位点在不同鹅群体中的分布情况。【结果】①经测序和拼接,获得鹅FREM1基因cDNA序列7 305bp,该序列包含一个完整的CDS(Coding Sequences)区,编码2 184个氨基酸。与四川白鹅和浙东白鹅相比,在豁眼鹅FREM1基因CDS序列上发现第4 515bp:T>C是错义突变,导致第1 505aa:Val>Ala变化,位于FREM1蛋白的CSPG重复结构域中第10个CSPG上。利用在线工具SIFT预测该氨基酸替换对蛋白功能的影响较小。通过I-Mutant ΔΔG和MUPro程序分析,p.1505V>A位点氨基酸替换大幅度降低了FREM1蛋白的稳定性。②FREM1基因在四川白鹅公、母鹅的眼睑和肾脏2个组织中均有表达,但肾脏表达水平远远高于眼睑,更为重要的是,公鹅FREM1基因的组织表达水平正好接近母鹅的2倍。ZHW(正常眼睑)和ZhW(上眼睑部分缺失)基因型鹅FREM1基因相对表达量无差异(P>0.05),虽然ZHZh(正常眼睑)基因型鹅的FREM1基因相对表达量是ZHW和ZhW基因型鹅的2倍多,但这可能是性别不同导致的差异。③豁眼鹅群体中基因型HH、Hh和hh的频率分别是0、0和1.0,等位基因H和h的频率分别是0和1.0,杂合度为0;四川白鹅群体中基因型HH、Hh和hh的频率分别是1.0、0和0,等位基因H和h的频率分别是1.0和0,杂合度为0;F1代群体中基因型HH、Hh和hh的频率分别是0、1.0和0,等位基因H和h的频率分别是0.5和0.5,杂合度为1.0。【结论】基因FREM1是决定鹅上眼睑性状的H基因座,该基因编码区1个纯合型错义突变导致了FREM1蛋白第10个CSPG结构域的变化,从而影响了FREM1蛋白的稳定性,基因FREM1的c.4514T>C突变可作为鹅豁眼性状重要的分子标记。
FREM1基因;豁眼性状;cDNA克隆;豁眼鹅;基因表达
0 引言
【研究意义】鹅豁眼性状呈伴性遗传,其形成主要受H和M两个基因座影响,其中H基因座位于Z染色体上,该基因座与眼睑发育有密切的关联[1]。从分子生物学角度研究鹅豁眼性状的遗传基础,阐释眼睑缺失表型形成的原因,对豁眼鹅遗传资源保护以及在肉鹅配套系中的利用方面具有重要的经济意义。此外,眼睑发育异常也出现于两类罕见的人类常染色体隐性遗传疾病Manitoba-oculo-tricho-anal(MOTA)综合征和Bifid nose and anorectal and renal anomalies(BNAR)综合征[2-4]。通过现代分子生物学手段,人们找到了一些与眼睑缺陷病症相关的染色体区域、关键通路和基因[5-10],但该病症的遗传机制尚未完全明了。因此,开展豁眼性状候选基因研究,既可阐明该性状的分子遗传机制,又为人类眼睑异常病症研究提供参考。【前人研究进展】ALAZAMI等[11]通过连锁分析,将MOTA综合征关联基因精细定位于人类染色体9p22.3,该区域FRAS-related extracellular matrix1(FREM1)基因与上眼睑发育相关[7]。基因FREM1编码基底膜的细胞外基质组份,与Fras1、Frem2共同组成胞外基质(extracellular matrix, ECM)蛋白, 从而调节胚胎发育过程中表皮基底膜和底层真皮之间的粘合;FREM1蛋白拥有典型的12个硫酸软骨素蛋白多糖(CSPG)重复结构域、氧化钙β(Cax-β)结构域和C型植物凝集素(C-type lectin)结构域,这些结构域在ECM蛋白家族都很保守,发挥着重要的生物学作用[12]。通过研究模型动物和人类MOTA综合征家族的基因FREM1序列信息,发现该基因的移码、外显子删除和错义突变均导致上眼睑残缺表型的出现[13-18]。【本研究切入点】检索GenBank和Ensemble等数据库,发现鸡、鸭、火鸡和斑胸草雀等禽类的FREM1基因均位于Z染色体上,而且像人类一样,FREM1 与CER1、ZDHHC21基因一起连锁[7],表明FREM1基因在各物种中保守性强,发挥的生物学作用比较相似。虽然鹅基因组的物理图谱尚未公布,但禽类基因组在很大程度上具有保守性和一致性[19-20],且禽类Z染色体上的基因排序非常相似[21-22],尤其鸡、鸭和鹅Z染色体上的基因及其位置顺序高度一致[23]。据此,假设基因FREM1是决定鹅上眼睑性状的H基因座,则与正常眼睑鹅相比,豁眼鹅基因FREM1编码区序列存在纯合型变异,使得鹅豁眼性状资源群中带有该变异信息的FREM1基因呈现Z染色体连锁模式。【拟解决的关键问题】本实验以鹅豁眼性状资源群为主要材料,克隆豁眼鹅和四川白鹅的FREM1基因,利用生物信息学手段分析该基因的结构和变异信息,结合其mRNA在不同基因型鹅组织中的表达情况,确证基因FREM1是豁眼形成的原因之一,为全面揭示眼睑性状的分子遗传机制奠定基础。
1 材料与方法
试验于2016年3—9月在辽宁省农业科学院创新中心分子实验室和辽宁省农业科学院彰武隆江牧业种鹅场进行。
1.1 试验动物和组织采集
从辽宁省农业科学院彰武隆江牧业种鹅场的鹅豁眼性状F2资源群中挑选出成年纯种豁眼鹅母鹅3只、四川白鹅6只(雌雄各半)、♂豁眼×♀四川白鹅F1代公鹅3只,颈动脉放血处死后,取其上眼睑和肾组织,立即投入液氮中,-80℃保存备用。
挑选出成年纯种豁眼鹅公鹅、四川白鹅公鹅和♂豁眼×♀四川白鹅正反交F1代公鹅各30只,翅静脉采血2mL,肝素抗凝并置于-20℃保存备用。
1.2 总RNA 的制备和反转录cDNA合成
鹅的上眼睑和肾组织,在液氮中研磨成粉末状,加入TRIzol裂解液裂解组织,用氯仿/异戊醇、异丙醇抽提,75%乙醇洗涤,适量无RNA 酶的水溶解。NanoDrop2000分光光度计测定总RNA的纯度和浓度,冻存于-80℃。
采用PrimeScript™ RT reagent Kit with gDNA Eraser进行cDNA反转录,实验操作按产品说明书进行。将所得的cDNA置于-20℃保存。
1.3 FREM1基因cDNA克隆与测序
根据GenBank已公布的鹅FREM1基因序列(XM_013193557)设计8 对引物(表1和图1),以合成的鹅cDNA为模板进行PCR扩增。PCR反应体系(25 μL):1.0μLcDNA、上下游引物各1.0 μL、2.5 μL dNTP Mix、0.25 μLKODFX、加ddH2O至25 μL。
反应条件:95°C 5 min;94°C 1min,53°C 1min,72°C 1.5min,35个循环;4℃保存。将扩增产物在1.0%琼脂糖凝胶中电泳,DNA磁珠法回收纯化试剂盒将PCR产物纯化,送北京华康同创生物技术有限公司测序。
1.4 鹅血红细胞基因组DNA提取
采用天根生化科技有限公司提供的血液基因组DNA提取试剂盒提取鹅基因组DNA。用1%琼脂糖检测提取DNA的质量。全波长紫外/可见光扫描分光光度计Nanovue检测DNA的浓度和纯度,并置于-20℃保存备用。

图1 RT-PCR扩增鹅基因FREM1片段所用引物示意图Fig.1 Schematic diagram of the primers used for amplication of the gene FREM1 by RT-PCR
1.5 PCR扩增与突变位点检测
根据GenBank已公布的鹅FREM1基因序列(Anser cygnoides domesticus breed Zhedong scaffold203_32, NCBI)设计引物(表1),以鹅总DNA为模板进行扩增。反应体系为:2×Taq PCRMaster Mix10μL,模板DNA2.0μL,引物F11的上下游引物各2.0μL,加灭菌蒸馏水至25μL。反应条件为:94℃预变性4min;94℃变性30s,60℃退火30s,72℃延伸2min,30个循环;72℃延伸10 min;4℃保存。1%的琼脂糖凝胶电泳检查PCR产物。PCR产物送北京华康同创生物技术有限公司进行直接测序。

表1 各引物基本信息Table 1 Descriptions of the 11 primers used in this study
1.6 qRT-PCR检测不同基因型鹅FREM1基因的表达水平
以梯度稀释的含有目的片段的质粒为标准品,做标准曲线,每个样品设置3个重复,对FREM1和ACTB进行实时定量PCR扩增。根据所获得的鹅FREM1基因序列设计定量PCR引物(表1)。反应体系(20 μL):模板cDNA 2.0 μL,2×Power SYBR® MasterMix 10.0 μL,上游引物0.5 μL,下游引物0.5 μL,加ddH2O至20 μL。反应条件:95℃ 30s;95℃ 5s,60℃ 40s(收集荧光), 45个循环;熔点曲线分析60—95℃。以鹅ACTB(XM_013174886)在肝脏组织中的表达作内标,通过Ct值与标准曲线,计算的目标基因的相对表达量和标准差,用 Excel 作图。
1.7 数据统计与分析
采用Primer 6.0和Beacon designer 7.8软件设计PCR、RT-PCR和qRT-PCR引物。获得的序列利用DNAMan软件、MEGA4.0和NCBI的Blastn等软件进行生物信息学分析。采用Excel和SPSS18.0软件处理试验数据、进行t检验、方差分析和显著性检验。
2 结果
2.1 鹅FREM1cDNA 克隆及生物信息学分析
以豁眼鹅和四川白鹅肾脏组织 cDNA 为模板,利用引物 F1-F8 (表1)进行 PCR 扩增。通过拼接多次RT-PCR序列获得了豁眼鹅和四川白鹅两个品种鹅的FREM1cDNA序列7 305 bp,该序列包含一个完整的CDS区,编码2 184个氨基酸。
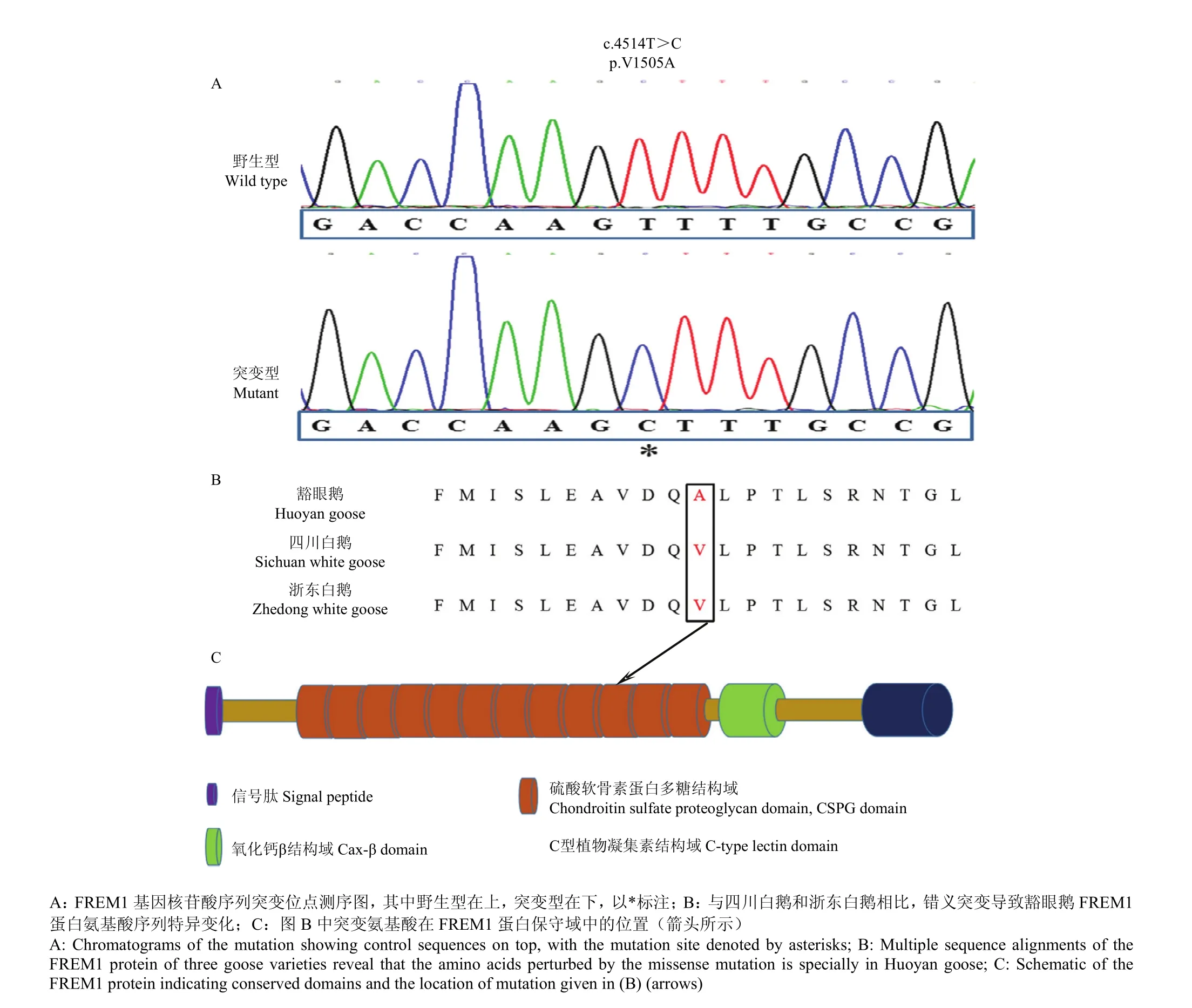
图2 基因FREM1突变导致鹅豁眼性状示意图。Fig. 2 Mutation in FREM1 is responsible for Huoyan trait of goose.
与正常眼睑表型的四川白鹅和浙东白鹅[24](XM_013193557.1)序列相比,在豁眼鹅FREM1基因CDS序列上发现10个单核苷酸突变,其中1个是错义突变,为第4 514 P:T>C(位于外显子24上)(图2-A),第1 505aa:缬氨酸(Val)>丙氨酸(Ala)变化(图2-B),位于FREM1蛋白的CSPG重复结构域中第10个CSPG上(图2-C)。利用在线工具SIFT(http://sift.jcv.org)预测氨基酸替代对蛋白功能的影响(预测阈值小于0.05则为有害)p.1505V>A位点预测值为0.51,表明该氨基酸替换对蛋白功能的影响较小[25]。由于检测到人类FREM1基因移码、外显子删除和错义突变等影响蛋白功能的变异情况,使得相应病例不但有眼睑缺失的表型,同时带有异常的发际线,隐眼、鼻裂、肾发育不全等症状,甚至有些不到成年就夭折[13-18],表明FREM1蛋白有重要的生物学功能,极其有害的突变会降低该等位基因携带者的适合度,因而,从另一个角度提示,豁眼鹅FREM1基因p.1505V>A的突变虽然可能是导致豁眼表型的原因之一,但是其影响的程度是比较温和的。
由于众多致病性的错义突变都会引起蛋白稳定性的变化,利用在线工具分析p.1505V>A位点氨基酸变化是否导致了FREM1蛋白稳定性的降低[26-27]。通过I-Mutant ΔΔG程序(http://gpcr2.biocomp. unibo.it/cgi/predictors/I-Mutant3.0/I-Mutant3.0.cgi;
Tem.25℃, pH 7.0)分析,结果显示,p.1505V>A位点的自由能变化为-1.95Kcal·mol-1,表明该氨基酸替换大幅度降低了FREM1蛋白的稳定性[26,28]。利用MUPro(http://mupro. proteomics.ics.uci.edu)在线工具分析也得到了同样的结果[29],其中,基于支持向量机(support vector machine,SVM)方法分析得分是-1,表明蛋白稳定性降低;基于神经网络(neural network,NW)方法分析得分是-0.95,表明蛋白稳定性降低。
2.2 鹅眼睑和肾脏组织FREM1基因mRNA表达水平分析
人类MOTA综合征表现为眼睑部分缺失,异常的发际线,隐眼、鼻裂、肾发育不全等[3-4],而基因FREM1也主要在人类和小鼠的表皮、眼部、肾脏和肺脏中表达[30],检测鹅基因FREM1在眼睑和肾脏组织中的表达情况,以验证该基因是否存在性别和不同表型之间的差别。
FREM1基因在四川白鹅公、母鹅的眼睑和肾脏2个组织中均有表达,但肾脏组织表达水平远远高于眼睑,更为重要的是,公鹅FREM1基因的组织表达正好是母鹅的2倍(图3),表明鹅基因FREM1表达可能与性别关联,即公鹅有2个FREM1基因,而母鹅只有1个。根据鹅FREM1基因变异和H基因座情况[1],定义c.4514T>C位点为H基因座重要的分子标记,其中,定义野生型(T)为H基因座的H等位基因,突变型(C)为H基因座的h等位基因。
根据鹅豁眼性状遗传模式分析[1],四川白鹅母鹅(正常眼睑)、豁眼鹅母鹅(上眼睑部分缺失)及其杂交F1代公鹅(正常眼睑)H基因座的基因型分别为ZHW、ZhW和ZHZh。图4显示了FREM1基因在不同基因型鹅眼睑组织中的相对表达情况。ZHW和ZhW基因型鹅的FREM1基因相对表达量无差异(P>0.05);ZHZh基因型鹅的FREM1基因相对表达量是ZHW和ZhW基因型鹅的2倍,这可能是不同性别鹅带有FREM1基因的数量不同所致(图3)。这些结果提示,FREM1基因两个外显子的错义突变可能并未影响该基因的表达,证实了豁眼鹅FREM1蛋白氨基酸序列变化可能只是影响了该蛋白稳定性的预测。
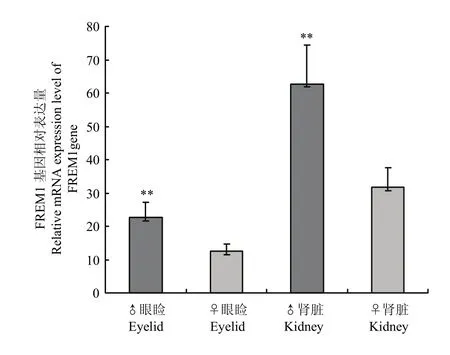
图3 FREM1基因在公母鹅眼睑和肾脏中的表达情况Fig. 3 Relative mRNA expression level of FREM1 gene in eyelid and kidney of male and female geese

图4 FREM1基因在不同基因型鹅的上眼睑组织中表达情况Fig. 4 Relative mRNA expression level of FREM1 gene in upper eyelid of different genotype of goose
2.3 鹅豁眼性状资源群基因FREM1变异及基因型频率情况
由于H座位呈伴性遗传,检测豁眼鹅、四川白鹅和其正反交F1群体中公鹅的基因型更具代表性。检测结果显示(表2),豁眼鹅H基因座上基因型HH、Hh和hh的频率分别是0、0和1.0,等位基因H和h的频率分别是0和1.0,杂合度为0;四川白鹅H基因座上基因型HH、Hh和hh的频率分别是1.0、0和0,等位基因H和h的频率分别是1.0和0,杂合度为0;F1代H基因座上基因型HH、Hh和hh的频率分别是0、1.0和0,等位基因H和h的频率分别是0.5和0.5,杂合度为1.0。结果表明,豁眼鹅群体是h等位基因纯合型,四川白鹅群体是H等位基因纯合型,这与鹅豁眼性状遗传模式分析的理论推理一致[1]。
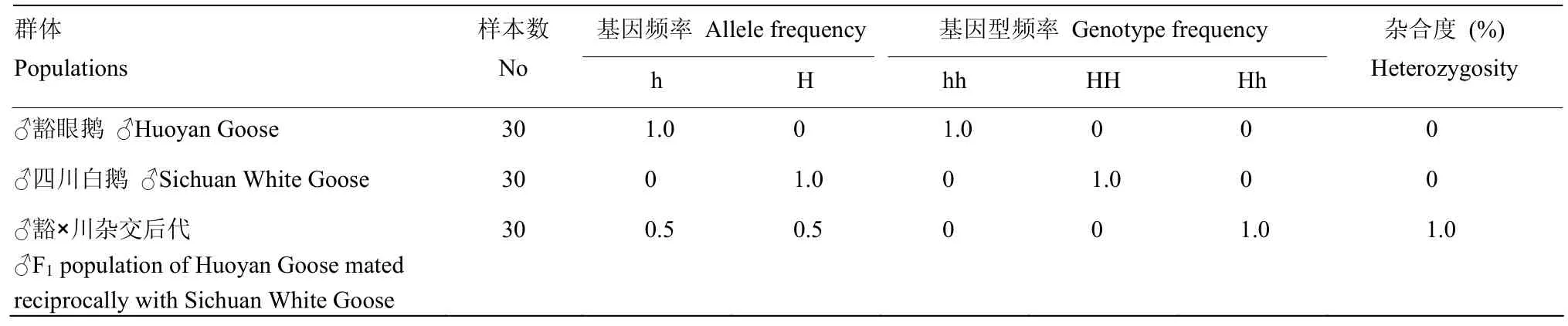
表2 鹅豁眼性状资源群H基因座变异情况Table 2 Current status of genetic variation at the H locus in F2resource population with Huoyan trait
3 讨论
本试验克隆了鹅FREM1基因的编码区序列,通过与正常眼睑鹅FREM1基因序列的比对分析,在豁眼鹅群体中发现纯合型错义突变c.4514T>C(图2-A),通过I-Mutant ΔΔG程序和MUPro在线工具分析,表明该突变位点对蛋白功能影响较小,可能只是导致了蛋白稳定性的降低,这需要进一步的验证。通过对FREM1基因在不同基因型鹅组织中的实时定量分析(图3和图4),表明鹅FREM1基因与Z染色体存在较强的关联性,同时,结合鹅豁眼性状资源群中基因FREM1变异分析(表2),笔者初步认为基因FREM1是鹅豁眼性状H座位的候选基因,其编码区c.4514T>C突变可作为鹅豁眼性状重要的分子标记。
FREM1蛋白拥有典型的12个CSPG重复结构域、Cax-β结构域和C-type lectin结构域,这些结构域在ECM蛋白家族都很保守,发挥着重要的生物学作用[12]。众多研究表明,带有CSPG结构域的蛋白家族与血小板衍生生长因子(platelet-derived growth factor)、成纤维细胞生长因子(fibroblast growth factor)、胶原蛋白V(collagen V)和胶原蛋白VI(collagen VI)等因子有互作效应[31-33],提示FREM1蛋白可能也有类似的作用[34]。豁眼鹅基因FREM1的氨基酸替换位于FREM1蛋白高度保守的CSPG重复结构域中第10个CSPG上(图2-C),通过在线工具预测,表明该位置的突变大大降低了蛋白的稳定性。由于豁眼性状是由H和M两个基因座互作而形成[1],推测豁眼鹅H基因座的FREM1氨基酸序列p.1505Val>Ala变化可能影响了与之有互作效应的因子,而该因子可能是M基因座[1],这为开展豁眼性状另一个常染色体上基因座研究指明了方向。
4 结论
基因FREM1是决定鹅上眼睑性状的H基因座,该基因编码区1个纯合型错义突变导致了FREM1蛋白第10CSPG结构域的变化,基因FREM1的c.4514T>C突变可作为鹅豁眼性状重要的分子标记。
[1] 于金成,李 喆,于 宁,赵 辉. 基于F2群体的豁眼鹅豁眼性状遗传分析. 中国农业科学, 2016, 49(19): 3845-3851.
YU J J, LI Z, YU N, ZHAO H. Genetic analysis of Huoyan trait based on F2resource population in Huoyan Goose. Scientia Agricultura Sinica, 2016, 49(19): 3845-3851.(in Chinese)
[2] LI A G, BAKIR M, HAMUD O A, GERAMI S. An autosomal recessive syndrome of nasal anomalies associatedwith renal and anorectal malformations. Clinical Dysmorphology, 2002, 11: 33-38.
[3] LI C, MARLES S L, GREENBERG C R, CHODIRKER B N, VAN DE KAMP J, SLAVOTINEK A, CHUDLEY A E. Manitoba Oculotrichoanal (MOTA) syndrome: report of eight new cases. American Journal of Medical Genetics Part A, 2007, 143(8):853-7.
[4] YEUNG A, AMOR D, SAVARIRAYAN R. Familial upper eyelid coloboma with ipsilateral anterior hairline abnormality: two new reports of MOTA syndrome. American Journal of Medical Genetics Part A, 2009, 149(4): 767-769.
[5] MINE N, IWAMOTO R, MEKADA E. HB-EGF promotes epithelial cell migration ineyelid development. Development, 2005, 132: 4317-4326.
[6] RAO W W Y, XIA Y, LIU C Y. Signaling pathways in morphogenesis of cornea and eyelid. The Ocular Surface, 2008, 6(1): 9-23.
[7] ALAZAMI A M, SHAHEEN R, ALZAHRANI F, SNAPE K, SAGGAR A, BRINKMANN B, BAVI P, LI A G, ALKURAYA F S. FREM1 mutations cause bifid nose, renal agenesis, and anorectal malformations syndrome. The American Journal of Human Genetics, 2009, 85(3):414-418.
[8] DAUWERSE J G, DIXON J, SELAND S. Mutations in genes encoding subunits of RNApolymerases I and III cause Treacher Collins syndrome. Nature Genetics, 2011, 43: 20-22.
[9] OHUCHI H. Wakayama Symposium: Epithelial-mesenchymal interactions in eyelid development. The Ocular Surface, 2012, 10(4): 212-216.
[10] SHI F, FAN Y, ZHANG L, MENG L, ZHI H, HU H, LIN A. The expression of Pax6 variants is subject to posttranscriptional regulation in the developing mouse eyelid. PloS One, 2013, 8(1):e53919.
[11] PETROU P, MAKRYGIANNIS A K, CHALEPAKIS G. The Fras1/Frem family of extracellular matrix proteins: structure, function, and association with Fraser syndrome and the mouse bleb phenotype. Connective Tissue Research, 2008, 49(3-4): 277-282.
[12] SMYTH I, DU X, TAYLOR M S, JUSTICE M J, BEUTLER B, JACKSON I J. The extracellular matrix gene Frem1 is essential for the normal adhesion of the embryonic epidermis. Proceedings of the National Academy of Sciences of the United States of America, 2004, 101(37):13560-13565.
[13] SLAVOTINEK A M, BARANZINI S E, SCHANZE D, LABELLEDUMAIS C, SHORT K M, CHAO R, YAHYAVI M, BIJLSMA E K, CHU C, MUSONE S, WHEATLEY A. Manitoba-oculo-tricho-anal (MOTA) syndrome is caused by mutations in FREM1. Journal of Medical Genetics, 2011, 48(6): 375-382.
[14] MITTER D, SCHANZE D, STERKER I, MÜLLER D, TILL H, ZENKER M. MOTA syndrome: Molecular genetic confirmation of the diagnosis in a newborn with previously unreported clinical features. Molecular Syndromology, 2012, 3(3):136-139.
[15] MATEO R K, JOHNSON R, LEHMANN O J. Evidence for additional FREM1 heterogeneity in Manitoba oculotrichoanal syndrome. Molecular Vision, 2012, 18: 1301-1311.
[16] WIRADJAJA F, COTTLE D L, JONES L, SMYTH I. Regulation of PDGFC signalling and extracellular matrix composition by FREM1 in mice. Disease Models and Mechanisms, 2013, 6(6):1426-1433.
[17] BECK T F, SHCHELOCHKOV O A, YU Z, KIM B J, HERNÁNDEZ-GARCÍA A, ZAVERI H P, BISHOP C, OVERBEEK P A, STOCKTON D W, JUSTICE M J, SCOTT D A. Novel frem1-related mouse phenotypes and evidence of genetic interactions with gata4 and slit3. PloS One, 2013, 8(3):e58830.
[18] NATHANSON J, SWARR DT, SINGER A, LIU M, CHINN A, JONES W, HURST J, KHALEK N, ZACKAI E, SLAVOTINEK A. Novel FREM1 mutations expand the phenotypic spectrum associated with manitoba-oculo-tricho-anal (MOTA) syndrome and bifid nose renal agenesis anorectal malformations (BNAR) syndrome. American Journal of Medical Genetics Part A, 2013, 161(3):473-478.
[19] GRIFFIN D K, ROBERTSON L B, TEMPEST H G, SKINNER B M. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenetic and Genome Research, 2007, 117(1-4):64-77.
[20] DAWSON D A, ÅKESSON M, BURKE T, PEMBERTON J M, SLATE J, HANSSON B. Gene order and recombination rate in homologous chromosome regions of the chicken and a passerine bird. Molecular Biology and Evolution, 2007, 24(7):1537-1552.
[21] NANDA I, SICK C, MÜNSTER U, KASPERS B, SCHARTL M, STAEHELI P, SCHMID M. Sex chromosome linkage of chicken and duck type I interferon genes: further evidence of evolutionary conservation of the Z chromosome in birds. Chromosoma, 1998, 107(3): 204-210.
[22] BACKSTRÖM N, BRANDSTRÖM M, GUSTAFSSON L, QVARNSTRÖM A, CHENG H, ELLEGREN H. Genetic mapping in a natural population of collared flycatchers (Ficedula albicollis): conserved synteny but gene order rearrangements on the avian Z chromosome. Genetics, 2006, 174(1):377-386.
[23] ISLAM F B, UNO Y, NUNOME M, NISHIMURA O, TARUI H, AGATA K, MATSUDA Y. Comparison of the chromosome structures between the chicken and three Anserid species, the Domestic Duck (Anas platyrhynchos), Muscovy Duck (Cairina moschata), and Chinese Goose (Anser cygnoides), and the delineation of their karyotype evolution by comparative chromosome mapping. TheJournal of Poultry Science, 2014, 51(1):1-3.
[24] LU L, CHEN Y, WANG Z, LI X, CHEN W, TAO Z, SHEN J, TIAN Y, WANG D, LI G, CHEN L. The goose genome sequence leads to insights into the evolution of waterfowl and susceptibility to fatty liver. Genome Biology, 2015, 16(1):1.
[25] KUMAR P, HENIKOFF S, NG P C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols, 2009, 4:1073-1081.
[26] YUE P, LI Z, MOULT J. Loss of protein structure stability as a major causative factor in monogenic disease. Journal of Molecular Biology, 2005, 353(2):459-473.
[27] KHAN S, VIHINEN M. Performance of protein stability predictors. Human Mutation, 2010, 31(6):675-684.
[28] CAPRIOTTI E, FARISELLI P, ROSSI I, CASADIO R. A three-state prediction of single point mutations on protein stability changes. BMC Bioinformatics, 2008, 9(2):1.
[29] CHENG J, RANDALL A, BALDI P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins: Structure, Function, and Bioinformatics, 2006, 62(4): 1125-1132.
[30] SHORT K, WIRADJAJA F, & SMYTH I. Let's stick together: the role of the Fras1 and Frem proteins in epidermal adhesion. IUBMB Life, 2007, 59(7): 427-435.
[31] BURG M A, TILLET E, TIMPL R, STALLCUP W B. Binding of the NG2 proteoglycan to type VI collagen and other extracellular matrix molecules. Journal of Biological Chemistry, 1996, 271(42): 26110-26116.
[32] GORETZKI L, BURG M A, GRAKO K A, STALLCUP W B. Highaffinity binding of basic fibroblast growth factor and platelet-derived growth factor-AA to the core protein of the NG2 proteoglycan. Journal of Biological Chemistry,1999, 274(24): 16831-16837.
[33] PETROU P, CHIOTAKI R, DALEZIOS Y, CHALEPAKIS G. Overlapping and divergent localization of Frem1 and Fras1 and its functional implications during mouse embryonic development. Experimental Cell Research,2007, 313(5):910-920.
[34] STAUB E, HINZMANN B, ROSENTHAL A. A novel repeat in the melanoma-associ ated chondroitin sulfate proteoglycan defines a new protein family. FEBS Letters,2002, 527(1-3):114-118.
(责任编辑 林鉴非)
Validation of Candidate Gene FREM1 of H Locus of Huoyan Trait in Goose
YU JinCheng, YU Ning, ZHAO Hui, LI Zhe
(Pratacultural & Animal Science Research Institute, Liaoning Academy of Agricultural Sciences, Shenyang 110161)
FREM1 gene; trait of upper eyelids colomobas; cloning of cDNA; Huoyan Goose; gene expression
2016-11-15;接受日期:2017-03-09
辽宁省博士启动基金资助项目(20141164)、国家水禽产业技术体系专项资金(CARS-43-23)、辽宁省科技型中小企业技术创新基金(2013-1)
联系方式:于金成,Tel:024-31029891;13898156386;E-mail:yujincheng_pi@126.com。通信作者赵辉,E-mail:zhaohui_sy@126.com。通信作者李喆,Tel:024-31029891;E-mail:acj423@163.com。
Abstract: 【Objective】Recessive Z-link inheritance of the Huoyan trait has been confirmed, but its genetic basis still has not been defined. Recessive mutations in the coding region of FRAS-related extracellular matrix1(FREM1) gene have been shown to cause partial or whole loss of upper eyelid in human and model mice. In this study, cloning, expression and polymorphism analysis of the goose FREM1 gene were carried out using goose resource populations with Huoyan trait as main materials, to verify the hypothesis that FREM1 is a candidate gene affecting eyelid traits of goose, which will lay a foundation for further study of molecular genetic mechanism of eyelid traits.【Method】The coding region of goose FREM1 gene was amplified and cloned from total RNA of goose upper eyelids and kidney tissues, which were collected from the Huoyan goose (3 females), Sichuan white goose(3males and 3females), F1population of male Huoyan goose mated with female Sichuan White goose(3males), byRT-PCR according to the full-length transcript of FREM1 (XM_013193557), and the bioinformation analysis was performed with it. Then, the expression of FREM1 gene in goose eyelid and kidney tissue was studied by real-time fluorescence quantitative PCR. The whole blood DNA was extracted from 30 adult male Huoyan geese, 30 male Sichuan white geese, and 30 male geese of F1population of Huoyan goose mated reciprocally with female Sichuan White goose, and the distribution of FREM1 gene mutations in different goose populations was detected by direct sequencing according to the genomic sequences of FREM1 (Anser cygnoides domesticus breed Zhedong scaffold203).【Result】①The 7 305bp cDNA sequences of FREM1 gene was obtained by sequencing and splicing, which contained a complete CDS region encoding 2 184 amino acids. Compared with the nucleotide sequences of the Sichuan White goose and Zhedong White goose, the c.4 515T> C missense mutation was found in CDS of FREM1 gene of Huoyan goose, which causes an Valine to Alanine change (p.Val1505Ala) at a highly conserved amino acid residue in FREM1’s tenth CSPG domain. Although, the p.1 505Val>Ala change was predicted to be “Tolerated” by SIFT, it was subsequently predicted to lead to decreased protein stability by the I-Mutant ΔΔG and MUPro program. ②FREM1 gene was expressed in the tissues of eyelids and kidneys of Sichuan White goose, but the expression level in kidney was much higher than eyelid, more importantly, the expression level of the male is nearly twice as that of the female. There was no significant difference in the relative expression of FREM1 gene between ZHW(Normal eyelids)and ZhW(Upper eyelids colomobas) genotypes (P>0.05). Although the relative expression of FREM1 gene of ZHZh(Normal eyelids) genotype was more than 2 times than that of ZHW and ZhW genotype, it could be caused by differences in gender. ③In Huoyan Goose populations, the frequencies of HH, Hh and hh genotypes were 0, 0 and 1.0, the frequencies of alleles H and h were 0 and 1.0, the heterozygosity was 0; The frequencies of HH, Hh and hh were 1.0, 0 and 0 in Sichuan white goose population, the frequencies of the alleles H and h were 1.0 and 0, the heterozygosity was 0 in Sichuan White Goose populations; The frequencies of HH, Hh and hh were 0, 1.0 and 0, the frequencies of the alleles H and h were 0.5 and 0.5, the heterozygosity was 1.0 in F1generation population of Huoyan Goose mated reciprocally with female Sichuan White Goose.【Conclusion】The FREM1 gene is the H locus which determines the eyelid traits of goose. A homozygous missense mutation in the coding region of the FREM1gene caused an Valine to Alanine change (p.Val1505Ala) at a highly conserved amino acid residue in FREM1’s tenth CSPG domain, which affected the stability of FREM1 protein. Therefore, mutation c.4514T> C of gene FREM1can be used as an important molecular marker.

