Small regulators making big impacts: regulation of neural stem cells by small non-coding RNAs
Small regulators making big impacts: regulation of neural stem cells by small non-coding RNAs
Neurodegeneration and traumatic brain injuries are leading causes of disability and present an enormous disease burden both in terms of patient suffering and healthcare cost. Treatment of brain lesions remains as a major challenge in medicine largely because of the limited regenerative capacity of the adult brain. Neural stem cells (NSCs), which are present in the embryonic nervous system and in specific regions of the adult brain, have the capacity for self-renewal by making identical daughter NSCs, and for differentiation into all neural cell types including neurons, astrocytes, and oligodendrocytes, therefore represent a promising therapeutic cellular source to treat brain lesions (Gage and Temple, 2013). However, NSCs are susceptible to genotoxic insults because of their proliferative nature, which could lead to unregulated NSC proliferation and brain tumors (Vescovi et al., 2006).
In order to harness the full medical potential of NSCs, it is critical to understand how NSCs precisely regulate proliferation, differentiation, and other basic biological features. MicroRNAs (miRNAs), which are ~22 nucleotide (nt) small non-coding RNAs regulating gene expression post-transcriptionally, have emerged as critical regulators of stem cells, tissue homeostasis, regeneration, and human diseases. In a recent study, Liu et al. (2017) provided novel insights into the critical roles of a sub-class of miRNAs, the canonical miRNAs, in the regulation of proliferation, lineage specification, gene expression, and cholesterol biogenesis of NSCs.
Canonical miRNAs are miRNAs that rely on both DROSHA-DGCR8 microprocessor and DICER for biogenesis. In the canonical miRNA biogenesis pathway, primary transcripts of miRNAs (pri-miRNAs) are transcribed by RNA polymerase II from miRNA genes or introns of protein-coding genes. Pri-miRNAs are first processed into pre-miRNAs by the microprocessor complex, which consists of the type III RNase DROSHA and the double-stranded RNA binding protein DGCR8. Pre-miRNAs are then processed into mature miRNAs by DICER, another type III RNase (Figure 1). Mature miRNAs are incorporated into the RNA-induced silencing complex (RISC) to suppress translation and/or destabilize mRNA targets (Ha and Kim, 2014).
In addition to converting pre-miRNAs into mature miRNAs, DICER is also required for the biogenesis of non-canonical miRNAs and endogenous small interfering RNAs (endo-siRNAs) (Cook and Blelloch, 2013). Non-canonical miRNAs are often originated from endogenous shRNAs or mirtrons, which have premiRNA-like structures generated directly from either transcription or RNA splicing. Endo-siRNAs are generated from DICER processing of endogenous long double-stranded RNAs (dsRNAs). Similar to canonical miRNAs, the non-canonical miRNAs and endo-siRNAs are also ~22 nt in length and can be loaded into RISC. Terefore, biogenesis of these small RNAs differs from canonical miRNAs in that only DICER is required rather than the DROSHA-DGCR8 microprocessor complex (Cook and Blelloch, 2013).
It has been demonstrated that DICER-dependent small non-coding RNAs play critical roles in the regulation of NSCs.Dicer-/-NSCs were successfully isolated from mouse embryonic brain (Andersson et al., 2010; Kawase-Koga et al., 2010). These NSCs can be cultured long termin vitrobut cannot differentiate into neurons or astrocytes. Because disruption of DICER completely abolishes biogenesis of canonical miRNAs, non-canonical miRNAs, and endo-siRNAs, it remains unclear which of the DICER-dependent small non-coding RNAs are responsible for the differentiation defects ofDicer-/-NSCs.
In a 2017 study by Liu and colleagues,Dgcr8-/-orDgcr8+/-NSCs were isolated from E13.5 mouse embryonic brains all from a single litter (Liu et al., 2017). Small RNA-seq analysis revealed thatDgcr8+/-NSCs abundantly express miRNAs such as miR-21, miR-30, and let-7 family members. Disruption ofDgcr8in NSCs led to an approximately 30-fold reduction of total miRNAs including all the abundantly expressed miRNAs, demonstrating that a great majority of the NSC-expressed miRNAs rely on the microprocessor for biogenesis. Interestingly, transcriptome analysis revealed that pri-miRNAs of many abundantly expressed miRNAs were present at significantly higher levels inDgcr8-/-than inDgcr8+/-NSCs. This agrees with the molecular function of the DROSHA-DGCR8 microprocessor to convert pri-miRNAs into pre-miRNAs (Figure 1). These data demonstrated that disruption ofDgcr8in NSCs led to a loss of canonical miRNAs.
Liu et al. (2017) further characterized self-renewal and differentiation ofDgcr8-/-NSCs. Similar toDicer-/-NSCs,Dgcr8-/-NSCs can be stably maintainedin vitrofor at least 20 passages. However, compared to controlDgcr8+/-NSCs,Dgcr8-/-NSCs proliferate at a much slower rate by a significantly extended G1 phase of the cell cycle, demonstrating that canonic miRNAs play an important role in promoting proliferation of undifferentiated NSCs. When cultured under permissive conditions,Dgcr8+/-NSCs efficiently differentiated into neurons, astrocytes, and oligodendrocytes as expected, whileDgcr8-/-NSCs underwent apoptosis under neuron- or oligodendrocyte-inducing conditions and failed to become astrocytes under astrocyte-inducing condition. These data demonstrated that among all the DICER-dependent small non-coding RNAs, canonical miRNAs are the key regulators for lineage specification of NSCs. Interestingly, previous studies demonstrated that canonical miRNAs are similarly required for the differentiation, but may be dispensable for self-renewal of pluripotent stem cells (PSCs) and dedifferentiation of somatic cells into induced pluripotent stem cells (iPSCs) (Wang et al., 2007; Liu et al., 2015). Whether canonical miRNAs are required by stem cells from other tissues for lineage specification, and how canonical miRNAs regulate lineage specification remain only partially understood.
Recently, Swahari et al. (2016) reported that disruption ofDicerorDgcr8in normal and cancerous mouse brain tissues induced DNA damage and resulted in increased apoptosis, suggesting inhibition of canonical miRNA biogenesis as a potential therapeutic target for brain tumors. Liu and colleagues confirmed the presence of excessive DNA damage, but failed to detect an increase of apoptosisinDgcr8-/-NSCs (Liu et al., 2017). Together with the findings thatDgcr8-/-NSCs undergo apoptosis under neuron- or oligodendrocyte-inducing conditions, these observations suggested that NSCs are more resistant to cell death caused by DNA damage when compared to differentiated neural cells. These findings raise the possibility that brain cancer stem cells, which likely originate from NSCs or immediate neural progenitor cells, are similarly resistant toDicerorDgcr8deficiency. In fact, it has been reported that cancer stem cells often exhibit increased resistance to radiation therapy or chemotoxic drugs (Vescovi et al., 2006). Terefore, it would be of great importance to further explore whether brain cancer stem cells exhibit resistance to the DNA damages induced by DICER or DGCR8 deficiency.
To better understandDgcr8deficiency on a molecular level, Liu and colleagues performed RNA-seq analysis to compare the expressional difference betweenDgcr8+/-andDgcr8-/-NSCs (Liu et al., 2017). Deletion ofDgcr8led to profound changes in the transcriptome of NSCs. Among the most affected transcripts, predicted mRNA targets of miR-21 and let-7s are significantly upregulated inDgcr8-/-NSCs. Gene set enrichment analysis (GSEA) further identified that genes associated with cell cycle progression, DNA replication, DNA damage repair, and neuronal system are significantly downregulated inDgcr8-/-NSCs. These data provided an explanation of whyDgcr8-/-NSCs are slower in proliferation, show increased DNA damage, and are defective in neuronaldifferentiation. However, it remains to be determined which canonical miRNAs regulate cell cycle progression, DNA damage repair, or lineage specification of NSCs. An important progress toward this goal was achieved recently in a study by Shenoy et al. (2015), in which let-7 and miR-125 were found to promote astrocyte differentiation inDgcr8-/-glial progenitor cells.
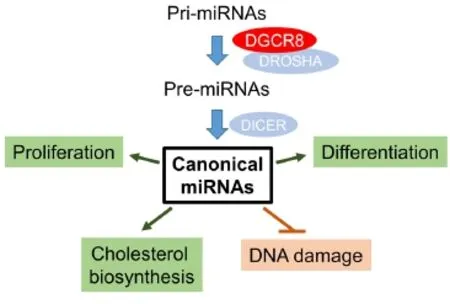
Figure 1 Canonical miRNAs are key regulators of neural stem cells.
Cholesterol is a key component of cell membrane and myelin sheath. Brain cholesterol is primarily producedde novoand is segregated from the rest of the body’s cholesterol sources by the blood-brain barrier. Mutations in cholesterol biosynthesis is the underlying cause of several human genetic diseases, such as Smith-Lemli-Opitz syndrome, Niemann-Pick Type C disease, and Conradi-Hunermann-Happle syndrome (Vance, 2012). Dysregulation of brain cholesterol balance is also closely associated with human neurodegenerative diseases such as Alzheimer’s disease and Parkinson’s disease (Vance, 2012). Rather unexpectedly, Liu et al. (2017) discovered that genes associated with cholesterol biosynthesis were significantly downregulated inDgcr8-/-NSCs. Expression ofSrebf1andSrebf2, the master transcriptional regulators for most enzymes required for cholesterol biosynthesis, was significantly downregulated and, importantly, levels of total cholesterol were reduced by approximately 30% inDgcr8-/-NSCs.
Consistent with this notion, Liu and colleagues discovered that genes associated with Alzheimer’s disease are significantly downregulated inDgcr8-/-NSCs (Liu et al., 2017). Taken together, these data not only demonstrate a previously under-appreciated role of canonical miRNAs in the regulation of cholesterol biosynthesis, they also suggest the canonical miRNA biogenesis pathway as a novel target to modulate brain cholesterol balance and to treat neurodegeneration. However, several questions have to be adequately addressed before this pathway can be exploited for cholesterol modulation.
First, how doesDgcr8deficiency lead to downregulation ofSrebf1andSrebf2? BecauseDgcr8inactivation will disrupt the biogenesis of all canonical miRNAs, it is likely that a negative transcriptional regulator ofSrebfgenes is de-repressed due to the lack of miRNAs. Identification of this putative negative regulator would allow identification of novel therapeutic targets and generate important insights into how miRNAs regulate brain cholesterol biosynthesis. Second, which miRNA or miRNA family is primarily responsible for the regulation of the genes associated with cholesterol biosynthesis? Inhibition of the canonical miRNA biogenesis pathway will affect expression of all miRNAs, which will in turn affect multiple functions of NSCs and the differentiated progenies. Identification of specific miRNAs that are responsible for cholesterol biosynthesis will allow targeting a limited number of miRNAs, which will be less toxic and more effective for pharmacological approaches to modulate brain cholesterol balance.
In summary, Liu and colleagues demonstrated that canonical miRNAs play critical roles in the regulation of proliferation, differentiation, DNA damage repair, and cholesterol biosynthesis in NSCs (Liu et al., 2017). In addition to the discoveries of basic mechanisms of NSC regulation, this study also suggested that future works aiming to understand how NSCs tolerate DNA damage could make the canonical miRNA biogenesis pathway a valid target to eliminate brain cancer stem cells and treat brain tumors. Similarly, an improved understanding of how canonical miRNAs regulate cholesterol biosynthesis would allow modulation of brain cholesterol balance and the treatment of neurodegenerative diseases.
Zhong Liu, Rui Zhao*
Department of Biochemistry and Molecular Genetics, Stem Cell Institute, University of Alabama at Birmingham, Birmingham, AL, USA
*Correspondence to:Rui Zhao, Ph.D., ruizhao@uab.edu.
Accepted:2017-03-10
orcid:0000-0003-3769-6550 (Rui Zhao)
Andersson T, Rahman S, Sansom SN, Alsio JM, Kaneda M, Smith J, O’Carroll D, Tarakhovsky A, Livesey FJ (2010) Reversible block of mouse neural stem cell differentiation in the absence of dicer and microRNAs. PLoS One 5:e13453.
Cook MS, Blelloch R (2013) Small RNAs in germline development. Curr Top Dev Biol 102:159-205.
Gage FH, Temple S (2013) Neural stem cells: generating and regenerating the brain. Neuron 80:588-601.
Ha M, Kim VN (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15:509-524.
Kawase-Koga Y, Low R, Otaegi G, Pollock A, Deng H, Eisenhaber F, Maurer-Stroh S, Sun T (2010) RNAase-III enzyme Dicer maintains signaling pathways for differentiation and survival in mouse cortical neural stem cells. J Cell Sci 123:586-594.
Liu Z, Skamagki M, Kim K, Zhao R (2015) Canonical microRNA activity facilitates but may be dispensable for transcription factor-mediated reprogramming. Stem Cell Rep 5:1119-1127.
Liu Z, Zhang C, Khodadadi-Jamayran A, Dang L, Han X, Kim K, Li H, Zhao R (2017) Canonical microRNAs enable differentiation, protect against DNA damage, and promote cholesterol biosynthesis in neural stem cells. Stem Cells Dev 26:177-188.
Shenoy A, Danial M, Blelloch RH (2015) Let-7 and miR-125 cooperate to prime progenitors for astrogliogenesis. EMBO J 34:1180-1194.
Swahari V, Nakamura A, Baran-Gale J, Garcia I, Crowther AJ, Sons R, Gershon TR, Hammond S, Sethupathy P, Deshmukh M (2016) Essential function of dicer in resolving DNA damage in the rapidly dividing cells of the developing and malignant cerebellum. Cell Rep 14:216-224.
Vance JE (2012) Dysregulation of cholesterol balance in the brain: contribution to neurodegenerative diseases. Dis Model Mech 5:746-755.
Vescovi AL, Galli R, Reynolds BA (2006) Brain tumour stem cells. Nat Rev Cancer 6:425-436.
Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R (2007) DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39:380-385.
10.4103/1673-5374.202938
How to cite this article:Liu Z, Zhao R (2017) Small regulators making big impacts: regulation of neural stem cells by small non-coding RNAs. Neural Regen Res 12(3):397-398.
Open access statement:This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
- 中国神经再生研究(英文版)的其它文章
- Anesthetic considerations for patients with acute cervical spinal cord injury
- Transplantation of autologous peripheral blood mononuclear cells in the subarachnoid space for amyotrophic lateral sclerosis: a safety analysis of 14 patients
- Anatomical distributional defects in mutant genes associated with dominant intermediate Charcot-Marie-Tooth disease type C in an adenovirusmediated mouse model
- Mechanisms responsible for the inhibitory effects of epothilone B on scar formation after spinal cord injury
- The mechanism of Naringin-enhanced remyelination after spinal cord injury
- Estrogen affects neuropathic pain through upregulating N-methyl-D-aspartate acid receptor 1 expression in the dorsal root ganglion of rats

