Viral Contamination Source in Clinical Microbiology Laboratory*
WANG Xin Ling, SONG Juan, SONG Qin Qin, YU Jie,LUO Xiao Nuan, WU Gui Zhen, and HAN Jun,#
Letter to the Editor
Viral Contamination Source in Clinical Microbiology Laboratory*
WANG Xin Ling1, SONG Juan1, SONG Qin Qin1, YU Jie1,LUO Xiao Nuan1, WU Gui Zhen2, and HAN Jun1,#
To understand the potential causes of laboratory-acquired infections and to provide possible solutions that would protect laboratory personnel, samples from a viral laboratory were screened to determine the main sources of contamination with six subtypes of Rhinovirus. Rhinovirus contamination was found in the gloves,cuffs of protective wear, inner surface of biological safety cabinet (BSC) windows, and trash handles. Remarkably, high contamination was found on the inner walls of the centrifuge and the inner surface of centrifuge tube casing in the rotor. Spilling infectious medium on the surface of centrifuge tubes was found to contribute to contamination of centrifuge surfaces. Exposure to sodium hypochlorite containing no less than 0.2 g/L available chlorine decontaminated the surface of the centrifuge tubes from Rhinovirus after 2 m in.
Laboratory-acquired infections (LAIs) are defined as those acquired through exposure to pathogens from laboratory or laboratory-related activities with or without symptoms. LAIs can lead to serious illness in laboratory personnel, which could be further transmitted to colleagues, family members, or others. In particular, LAIs contribute to the unintentional release of a significant amount of pathogenic organisms are into the environment,resulting in immediate or long-term risk to humans,animals, and the environment. Worldwide reports of LAIs have been published for the last 100 years. In a large LAI survey by Pike in 1976, among the 3921 cases examined, 164 deaths were attributed to LAIs[1]. Although laboratory safety measures have been published and training and education about these measures have been provided, exposure and LAIs, including those due to Brucella, Salmonella,Neisseria, and Orthopoxvirus are still reported[2-5]. Our previous study demonstrated that contamination with live pathogens also occurred during tissue homogenization either by ultrasonic processor or by tissue disperser[6]. This suggests that laboratory-acquired diseases pose an occupational hazard to clinical microbiologists. Thus, risk identification of experimental activities is important to prevent the aforementioned infections in personnel working in microbiology laboratories.
In this study, to gather risk information on viral contamination in clinical laboratories, a study on viral contamination source was conducted with human rhinovirus (HRV) as a model. Six subtypes of HRV (100 × TCID50) were evaluated. Specimens were collected from personal protective equipment (PPE)and surface of laboratory instruments during experimentation for two months. The experimental activities involved viral propagation, titration,collection, centrifugation, and aliquoting. After experimentation, many sites were sampled monthly to detect the presence of HRV RNA. All specimens were collected and sampled at least tw ice from the same laboratory. Using quantitative real-time polymerase chain reaction (qRT-PCR), we were unable to detect any HRV-positive specimens in the first month, excluding gloves and cuffs of protective clothes (Table 1). After the second month,HRV-positive samples were detected among those collected from the inner wall of the centrifuge (4/8),centrifuge tube casing in the rotor (7/24), inner surface of the biological safety cabinet (BSC)windows (1/5), and trash handles (1/4) (Table 2). The results showed that infectious material can easily spill or splash onto the gloves and cuffs of protective clothes in BSC indicating that they are one of the main sources of viral contamination in microbiology laboratories. Thus, we suggest that gloves should bechanged as frequently as possible and before leaving the BSC and laboratory. We also suggest that an oversleeve should be used to cover the cuffs of protective clothes, which can be removed before leaving the BSC.
Remarkably, higher count of HRV was noted on the inner wall of centrifuge tube casing in the rotor,while the inner surface of the BSC windows and trash handles showed weak HRV contamination(Table 2). This suggests that many LAIs due to low-risk organisms remain unidentified. Therefore,identification of contamination sites and prediction of pathogen contamination in laboratories are very important to alert personnel and avoid LAIs. Additionally, the risk of contamination increases withthe time of experimentation using centrifuges, BSCs,and incubators (Table 2). The inner surface of the BSC window, inner wall of centrifuge, and centrifuge tube casing were HRV-positive in the second round of sampling, indicating accumulation of contamination over time. The risk of infection to laboratory personnel increases when viral experiments are performed without implementing proper disinfection techniques. Thus, our results suggest that decontamination is necessary and should be performed thoroughly in viral laboratories at least once a month.
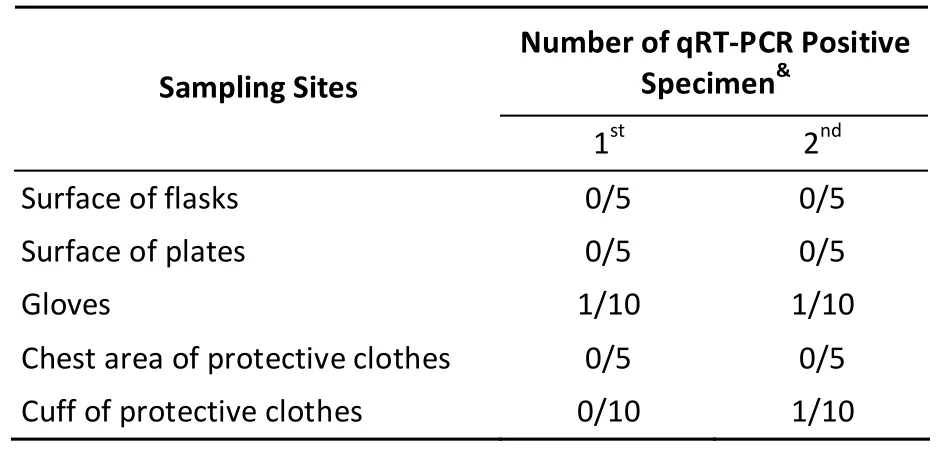
Table 1. Results of Surface Sampling of Disposable Materials in the HRV Laboratory
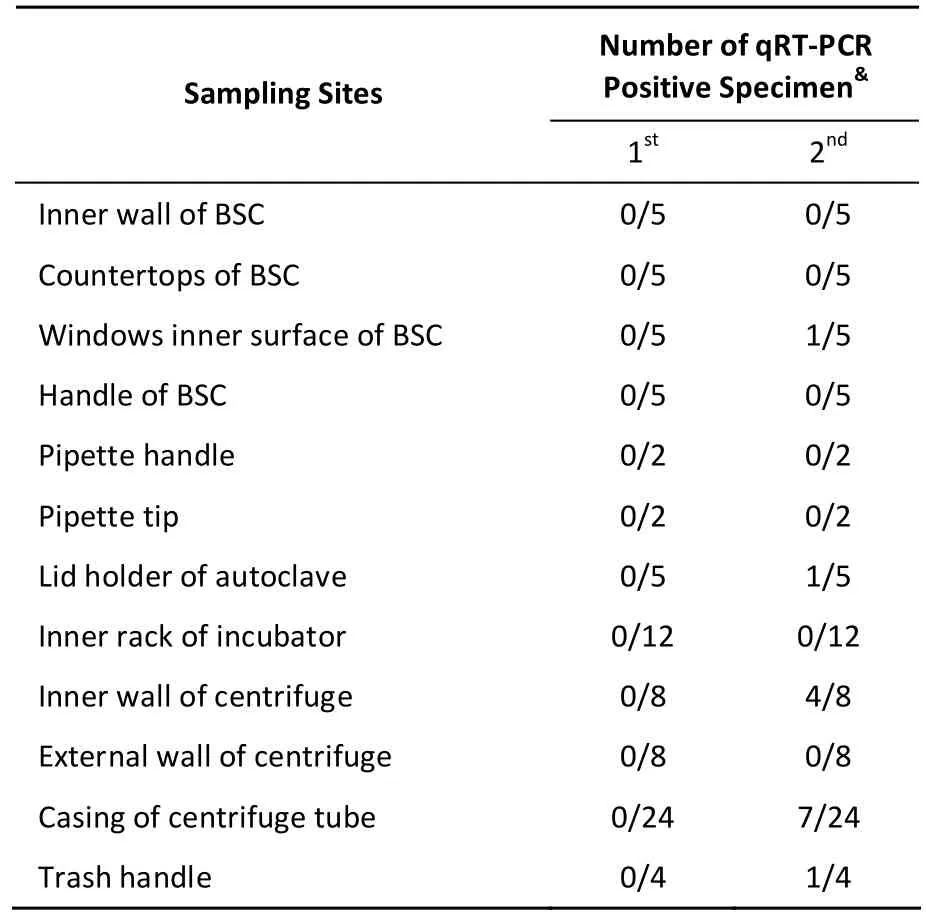
Table 2. Results of Surface Sampling of Instruments in the HRV Laboratory
Centrifugation is one of the most important steps during microbiological experiments. Therefore,in viral laboratories, the risk of contamination is the highest for centrifuge surfaces, and this can spread infectious particles and produce aerosols that pose a threat to laboratory personnel. To determine possible sources of contamination, HRV-18 was used to simulate the activities of viral preparation after the laboratory was decontaminated. After centrifugation, HRV contamination was detected in samples collected from both the rotor casing and inner walls of the centrifuge (Table 3). Our results also showed that during centrifugation, presence of high volumes of liquid likely leads to spilling and splashing of infectious materials on to the surfaces of the tubes, tube casing in the rotor, and inner wall of the centrifuge. The frequency of HRV-positive samples detected in the casing of 2 m L centrifuge tubes containing 1.5 m L, 1.8 m L, and 2.0 m L of viral supernatant was 1/24, 3/24, and 2/24, respectively(Table 3), while that of HRV-positive samples detected on the inner wall of centrifuge was 1/8. HRV nucleotides were found in the open and outer wall of centrifuge tubes, which contained 1.8 m L(2/72) and 2.0 m L (2/72) viral supernatant. No HRV nucleotides were detected in the 1.5 m L group(Table 3). This suggested that the outer surfaces of the centrifuge tubes are more likely to be contam inated during viral preparation, which can then lead to contamination of both the tube casing in the rotor and inner wall of the centrifuge. To prevent splashing and spilling during centrifugation,the centrifuge tubes should be tightly closed,high-quality tubes should be used, the volume of viral supernatant should not be more than 2/3 of the total volume of the tube, and the surface of centrifuge tubes should be decontaminated before centrifugation.
We next assessed the effectiveness of sodium hypochlorite in elim inating HRV contam inates fromthe surface of tubes. After 20 μL 100 × TCID50HRV was placed on the surface of tubes, gauze containing sodium hypochlorite (0.5 g/L, 0.25 g/L, 0.2 g/L, and 0.1 g/L available chlorine) was used to cover the surface of tubes for 2 m in, 10 m in, 20 m in, and 30 m in. No HRV nucleotides were detected on the surface of tubes covered with sodium hypochlorite solution (0.5 g/L, 0.25 g/L, and 0.2 g/L available chlorine) for no less than 2 m in. Our results demonstrated that 0.05-0.5 g/L available chlorine could effectively eliminate HRV from the surface of centrifuge tubes.
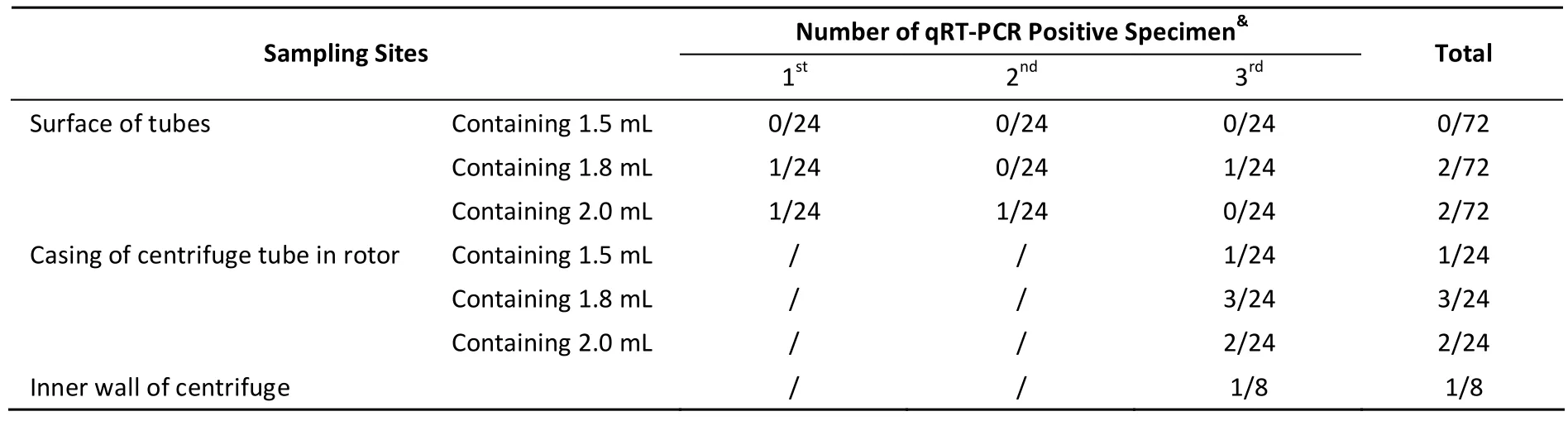
Table 3. Results of Surface Sampling of Centrifuge and Centrifuge Tubes in the HRV Laboratory
#Correspondence should be addressed to: HAN Jun,E-mail: hanjun_sci@163.com
Biographical note of the first author: WANG Xin Ling female, born in 1991, Bachelor, majoring in medical virology.
Accepted: August 1, 2016
REFERENCES
1. Pike RM. Laboratory-associated infections: summary and analysis of 3921 cases. Health Lab Sci, 1976; 13, 105-14.
2. Horvat RT, El Atrouni W, Hammoud K, et al. Ribosomal RNA sequence analysis of Brucella infection misidentified as Ochrobactrum anthropi infection. J Clin Microbiol, 2011; 49,1165-8.
3. Sejvar JJ, Johnson D, Popovic T, et al. Assessing the risk of laboratory-acquired meningococcal disease. J Clin M icrobiol,2005; 43, 4811-4.
4. Singh K. It's time for a centralized registry of laboratory-acquired infections. Nat Med, 2011; 17, 919.
5. Wei Q, Jiang MN, Han J, et al. Immune Control Strategies for Vaccinia Virus-related Laboratory-acquired infections. Biomed Environ Sci, 2014; 27, 142-6.
6. Song J, Zhou W, Wang Y, et al. Contam ination of live virus during tissue homogenizing by ultrasonic processor and tissue disperser. Biomed Environ Sci, 2012; 25, 167-71.
10.3967/bes2016.082
March 26, 2016;
*This work was supported by the China Mega-Project for Infectious Disease (2011ZX10004-001, 2012ZX10004401,2012ZX10004215, and 2013ZX10004805002); and the SKLID Development Grant (2011SKLID104).
1. State Key Laboratory for Infectious Disease Prevention and Control, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing 102206, China; 2. National Institute for Viral Disease Control and Prevention,Chinese Center for Disease Control and Prevention, Beijing 102206, China
 Biomedical and Environmental Sciences2016年8期
Biomedical and Environmental Sciences2016年8期
- Biomedical and Environmental Sciences的其它文章
- Circulating MicroRNAs as Novel Diagnostic Biomarkers for Very Early-onset (≤40 years) Coronary Artery Disease*
- Structural Modulation of Gut Microbiota in Rats with Allergic Bronchial Asthma Treated with Recuperating Lung Decoction*
- Expression of Peroxiredoxins and Pulmonary Surfactant Protein A Induced by Silica in Rat Lung Tissue*
- Distribution Characteristics of Spermophilus dauricus in Manchuria City in China in 2015 th rough ‘3S' Techno logy*
- Alcohol Drinking, Dyslipidemia, and Diabetes: A Population-based Prospective Cohort Study among lnner Mongolians in China*
- Bio logical Effec ts o f Clo th Con taining Specific Ore Pow der in Patien ts w ith Po llen Allergy
