Identification of Avian-Derived Ingredients in Livestock and Poultry Meat by PCR Technology
, , , , , , ,
Institute of Poultry, Chinese Academy of Agricultural Sciences, Yangzhou 225125, China
1 Introduction
Fake meat or meat adulteration has led to a serious violation of the legitimate rights and interests of consumers, and has directly affected the health of consumers, posing an important challenge to the current food quality control. Therefore, it is particularly important to test whether there is adulteration in the raw meat for food adulteration, and the key point is to quickly and accurately identify the source of meat ingredients[1]. PCR technology has characteristics of strong specificity, high sensitivity, easy operation and high efficiency, so it has become one of the most commonly used methods to identify the meat products[2]. Currently, because of its unique advantages, PCR technology has been widely used in the domestic and international study of adulterated meat, and there have been innovative results[3-5]. In terms of target gene selection, the animal’s mitochondrial genome DNA sequence has high degree species-specificity, and it is the primary target site for designing qualitative detection of meat ingredients, including cytochrome b (Cyt b) gene, 12S rRNA, 16S rRNA, D-Loop geneetc.[6-8]. In the West, there was once detection on nearly a thousand meat products, and the results showed that the mark of nearly 20% of the products was not exactly in line with the varieties[9]. Therefore, it is necessary to establish rapid and accurate detection method, study the animal-derived food safety testing technology, and identify the animal-derived ingredients in foods, which is conducive to safeguarding the interests of consumers. The technology based on polymerase chain reaction (PCR) has gradually become the core method to identify meat types. According to the difference in site for gene sequence of different species, many scholars designed specific primers and used PCR reaction to achieve exponential amplification of characteristic gene fragments in food, and then identified the possible sources of species in food by electrophoresis. Based on mitochondrial 12S rRNA gene, Girishetal. (2005)[10]used PCR-RFLP to successfully identify beef, buffalo meat, sheep meat and goat meat. Based on mitochondrial 12S rRNA and 16S rRNA gene, Ghovvatietal. (2009)[11]used multiplex PCR technique to identify ruminants, poultry and pigs. Soaresetal. (2010)[12]used double PCR to successfully detect poultry meat in pork. Chen Wenbingetal. (2005)[13]used single PCR and double PCR to identify the pork, beef, mutton, chicken and other meat ingredients in food. With yak, cattle, buffalo as research materials, Chen Dongetal. (2008)[14]found three specific enzyme cutting sites in the universal primer amplified fragment area of bovine mitochondrial 12SrRNA gene, which could be used for identification of bovine species sources of mixed fresh beef products. Based on the difference in site of animal mitochondrial cytochrome b gene, He Weilingetal. (2012)[15]designed two groups of five multiplex PCR primers of different length, established and optimized multiplex PCR reaction system, and used electrophoresis to amplify the differences in the molecular weight so as to realize rapid identification of four kinds of meat (pork, beef, mutton and chicken). With swine mitochondrial 12S rRNA gene sequence as the target site, Wang Yingetal. (2013)[16]designed primers and probes, conducted fluorescence quantitative PCR amplification, and established pig-derived ingredient detection method. In this study, with 16S rRNA gene sequence as the target gene, we performed the gene sequence alignment, and designed specific primers of chicken, pigeon and quail. With DNA of mutton, beef, pork, rabbit, pigeon, quail, chicken, duck and goose as template, we established the rapid detection method to identify chicken, pigeon and quail by PCR technology.
2 Materials and methods
2.1MaterialsWith the commercially available qualified mutton, beef, pork, rabbit, pigeon, quail, chicken, duck and goose as materials, each animal sample was thoroughly minced, mixed, and stored at -20℃ for later use.
2.2Methods
2.2.1DNA extraction. The centrifugal column genomic DNA extraction kit (Beijing Tiangen Biotech Co., Ltd.) was used to extract total DNA. The total DNA sample extracted was dissolved in 100 ul TE eluent, and stored at -20℃.
2.2.2Primer design and synthesis. According to 16S rDNA sequence, the Primer Premier5.0 software was used to select specific primer pairs for different species, and they were synthesized by Sangon Biotech (Shanghai) Co., Ltd. The right amount of primer was taken, dissolved in ultrapure water after autoclaving, and made into 10 umol/L stock solution. Primer sequence, Tm value and amplified fragment size can be shown in Table 1.
Table1Primersequence,TmvalueandPCRproductsize
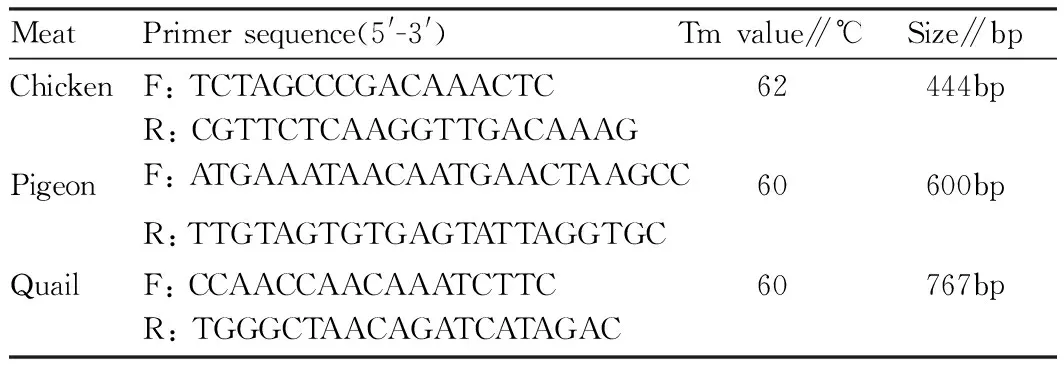
MeatPrimersequence(5'-3')Tmvalue∥℃Size∥bpChickenF:TCTAGCCCGACAAACTCR:CGTTCTCAAGGTTGACAAAG62444bpPigeonF:ATGAAATAACAATGAACTAAGCCR:TTGTAGTGTGAGTATTAGGTGC60600bpQuailF:CCAACCAACAAATCTTCR:TGGGCTAACAGATCATAGAC60767bp
2.2.3PCR amplification and detection. PCR amplification system (25 μL): 12.5 ul of 2×Tag Master Mix (Nanjing Boerdi Biotech Co., Ltd.), 0.5 μL of 10 μmol/L primer, 1 μL of template, 10.5 μL of sterile water. Reaction program: 95℃ 5 min, (95℃ 30 s, 60-62℃ 30 s, 72℃ 60 s) 28 cycles, 72℃ 4 min. After completion of the reaction, the PCR product was taken out, and 1.5% low melting point agarose (Promega Corporation) gel electrophoresis was used for PCR product detection.
2.2.4PCR product sequencing. After electrophoresis, the PCR products were recovered and purified and sent to Invitrogen Trading (Shanghai) Co., Ltd. for sequencing, and the bidirectional sequencing was used for alignment and verification. The sequencing results were compared with the known sequences listed in GenBank.
3 Results and analysis
3.1PCRtestresultsofdifferentanimalsThree pairs of primers were designed for each animal separately, and through optimization of reaction conditions, the same primer was used for PCR amplification of the nine animals selected. Finally, specific primers were selected from each animal for 1.5% agarose gel electrophoresis (Fig. 1). The PCR products were recovered, purified and sequenced, and the sequencing results were compared with the known sequences listed in GenBank. The results showed that the homology was more than 99% between chicken sequencing results and the published sequences (GenBank accession No. of AB086102.1, GU261713.1, GU261678.1), between pigeon sequencing results and the published sequences (GenBank accession No. of X87858.1, KP258178.1), between quail sequencing results and the published sequences (GenBank accession No. of AF302070.1, AB073301.1), basically in line with expectation. They were identified as chicken, pigeon and quail ingredients, respectively.
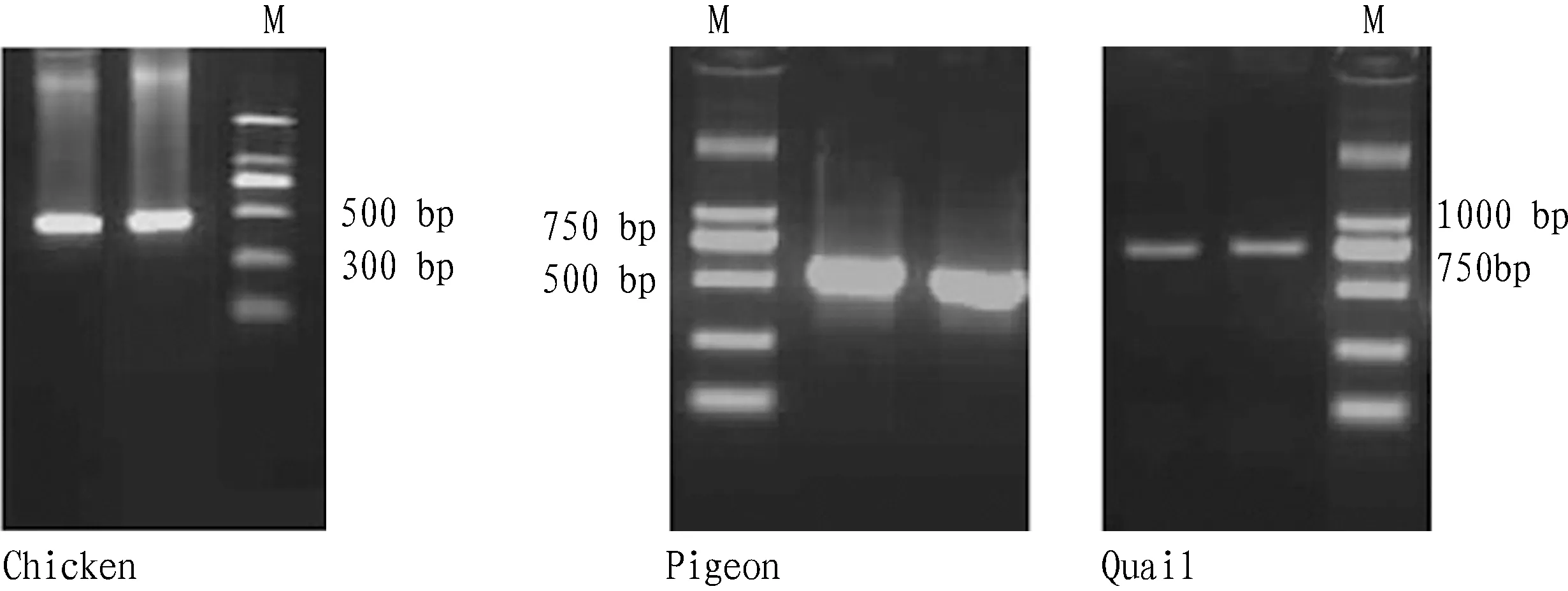
M:DL 2000 DNA marker
Fig.1PCRproducts

1. blank; 2. positive control; 3. mutton; 4. beef; 5. pork; 6. rabbit; 7. pigeon; 8. quail; 9. chicken; 10. duck; 11. goose; 12. DL 2000 DNA marker
Fig.2PCRamplificationresults
3.2SpecificityoftheprimerWith DNA of mutton, beef, pork, rabbit, pigeon, quail, chicken, duck and goose as template, we studied the specificity of chicken, pigeon and quail primers, respectively (Fig. 2). The positive control in Fig. 2 included chicken, pigeon and quail. Using the chicken primer, only chicken DNA template could be amplified into 444 bp target band; using the pigeon primer, only pigeon DNA template could be amplified into 600 bp target band; using the quail primer, only quail DNA template could be amplified into 767 bp target band.
4 Discussions
In this study, the specific primers of different species were designed according to the site difference in 16s RNA sequence of chicken, pigeon and quail mitochondrial genome, for PCR amplification and detection. After repeated tests, a quick and accurate chicken, pigeon and quail PCR identification method was established, with less sample consumption, simple reaction system and low cost. In the next step, we will carry out the 16s RNA sequence RTFQ PCR technology, further develop different methods for analysis of animal-derived ingredients in foods, explore new ways for meat quality and safety control, provide technical support to protect the interests of consumers, and provide technical supervision to ensure healthy development of the meat industry.
[1] HE WL, HUANG M, ZHANG C. Recent technological advances for identification of meat species in food products[J]. Food Science, 2012,33(3):304-307. (in Chinese).
[2] BALLIN NZ, VOGENSEN FK, KARLSSON AH. Species determination-can we detect and quantify meat adulteration[J]. Meat Science, 2009, 83(2): 165-174.
[3] YIN RH, BAI WL, WANG JM,etal. Development of an assay for rapid identi cation of meat from yak and cattle using polymerase chain reaction technique[J]. Meat Science, 2009, 83(1): 38-44.
[4] GHOVVATI S, NASSIRI MR, MIRHOSEINI SZ,etal. Fraud identi cation in industrial meat products by multiplex PCR assay[J]. Food Control, 2009, 20(8): 696-699.
[5] CHEN D, BAI F, ZHOU ML,etal.Differentiation of Bos grunniens, Bos Taurus, and Bubalus from meat products mixture based on mitochondrion 12S rRNA gene[J]. Hereditas, 2008, 30(8):1008-1014. (in Chinese).
[6] GIRISH PS, ANJANEYULU ASR, VISWAS KN,etal. Meat species identi cation by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) of mitochondrial 12S rRNA gene[J]. Meat Science, 2005, 70(1): 107-112.
[7] MURUGAIAH C, NOOR ZM, MASTAKIM M,etal. Meat species identification and Halal authentication analysis using mitochondrial DNA[J]. Meat Science, 2009, 83(1): 57-61.
[8] MANE BG, MENDIRATTA SK, TIWARI AK. Polymerase chain reaction assay for identi cation of chicken in meat and meat products[J]. Food Chemistry, 2009,116 (3): 806-810.
[9] BALLIN NZ, VOGENSEN FK, KARLSSON AH. Species determination: can we detect and quantify meat adulteration[J]. Meat Science, 2009, 83(2): 165-174.
[10] GIRISH PS, ANJANEYULU ASR, VISWAS KN,etal. Meat species identi cation by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) of mitochondrial 12S rRNA gene[J]. Meat Science, 2005, 70(1): 107-112.
[11] GHOVVATI S, NASSIRI MR, MIRHOSEINI SZ,etal. Fraud identi cation in industrial meat products by multiplex PCR assay[J]. Food Control, 2009, 20(8): 696-699.
[12] SOARES S, AMARAL JS, MAFRA I,etal. Quantitative detection of poultry meat adulteration with pork by a duplex PCR assay[J]. Meat Science, 2010, 85(3): 531-536.
[13] CHEN WB, SHAO BY, LIAO XB,etal. Application of PCR detection for some animal components in processed food[J]. Food Science, 2005, 26(8): 338-342. (in Chinese).
[14] CHEN D, BAI F, ZHOU ML,etal. Differentiation of Bos grunniens, Bos Taurus, and Bubalus from meat products mixture based on mitochondrion 12S rRNA gene[J]. Hereditas,2008,30(8):1008-1014. (in Chinese).
[15] HE WL, ZHANG C, YANG J,etal. A quick multiplex PCR method for the identification of four meat ingredients in food products[J]. Scientia Agricultura Sinica, 2012, 45(9):1873-1880. (in Chinese).
[16] WANG Y, SHI YY, LIU JH,etal. Detection of porcine-derived materials in meat products by real time PCR method[J]. Journal of Food Safety and Quality, 2013,4(5):1529-1534. (in Chinese).
 Asian Agricultural Research2016年12期
Asian Agricultural Research2016年12期
- Asian Agricultural Research的其它文章
- Shanghai "Maker" Urban Culture Construction in the Context of Big Cloud Movement
- Realistic Meaning of Agricultural Crisis Theory of Marx and Engels
- Current Situation and Mental Health and Intervention Measures of Left-behind Children
- Analysis on Development of Rural Financial Management Informationization
- Survival Predicament and Recommendations for Second Generation Farmers in Northwest China
- Problems in Development of Rural E-commerce and Logistics and Recommendations
