甜荞木瓜类半胱氨酸蛋白酶基因FeRD21的克隆与表达分析
*通信作者:刘志雄,博士,副教授,主要从事植物发育遗传学、育种与良种繁育研究。 E-mail:zxliu77@yahoo.com
甜荞木瓜类半胱氨酸蛋白酶基因FeRD21的克隆与表达分析
方正武1,李来运2,李晓方1,刘志雄1,2*
(1 长江大学 作物遗传育种研究所暨农作物多基因型种群育种技术中心,湖北荆州 434025;2 长江大学 园艺园林学院,湖北荆州 434025)
摘要:该研究以旱区小杂粮作物甜荞(Fagopyrum esculentum)为材料,采用同源克隆、RACE技术和实时荧光定量RT-PCR方法,对其半胱氨酸蛋白酶基因(FeRD21)进行了分离和表达分析。结果表明:(1)FeRD21基因cDNA全长1 750 bp,包含1个1 407 bp的完整开放阅读框,编码468个氨基酸。(2)蛋白序列比对发现,甜荞FeRD21全酶包括信号肽、N末端自主抑制前体区域、蛋白酶、脯氨酸富含结构域和C末端颗粒体蛋白结构域,同时,其蛋白酶结构域包含1个木瓜类蛋白酶家族保守的催化三连体活性位点:Cys168-His304-Asn324。(3)分子系统发生分析证实,其与拟南芥的RD21一致性最高,属类RD21半胱氨酸蛋白酶类。(4)基因表达分析表明,FeRD21能被干旱、高盐、ABA和衰老胁迫诱导。
关键词:非生物胁迫;甜荞;木瓜类半胱氨酸蛋白酶;FeRD21
收稿日期:2014-11-25;修改稿收到日期:2015-01-07
基金项目:国家自然科学基金(31101202);国家公益性行业(农业)科研专项(201303008);湖北省重点(优势)学科作物学(长江大学)
作者简介:方正武(1977-),男,博士,讲师,主要从事作物遗传育种研究。E-mail:fangzhengwu88@163.com
中图分类号:Q786 文献标志码:A
Clone and Expression Analysis of a Papain-like Cysteine
Protease Gene(FeRD21) inFagopyrumesculentum
FANG Zhengwu1,LI Laiyun2,LI Xiaofang1,LIU Zhixiong1,2*
(1 Institute of Crop Genetics and Breeding,Yangtze University,Jingzhou,Hubei 434025,China;2 College of Horticulture and Gardening,Yangtze University,Jingzhou,Hubei 434025,China)
Abstract:Fagopyrum esculentum(buckwheat,Polygonaceae) is a multi-food-use pseudocereal with healing benefits and is growing on arid areas.(1)Based on homology and RACE method,a RD21 orthologous gene from buckwheat was isolated and identified.The RD21 homologous gene from F.esculentum transcript was 1 750 bp and contained a 1 407 bp ORF(Open Reading Frame,ORF) encoding 468 amino acids.(2)Protein sequence alignment and phylogenetic analyses grouped FeRD21 into PLCPs subfamily members which carry a C-terminal granulindomain.(3)The protease of FeRD21 was highly conserved and harbored the conservation sites of catalytic residues Cys168-His304-Asn324.(4)Expression analysis suggested that FeRD21 was up-regulated by salt,dehydration,ABA,and senescent treatments,which showed adifferent way in response to stresses with RD21 in Arabidopsis.Our results indicated that FeRD21 might be involved the stress-responsive pathways in F.esculentum.
Key words:abiotic stresses;buckwheat;papain-like cys protease;FeRD21
Papain-like Cys proteases(PLCPs) are a large class of proteolytic enzymes associated withdevelopment,immunity,stress tolerance and senescence[1-3].PLCPs show the typical papain-like fold of twodomains,an α-helix-richdomain and a β-barrel-likedomain,separating a substrate-binding grove containing the catalytic triad Cys-His-Asn[4-5].Moreover,PLCPs are produced with a N-terminal auto-inhibitorydomain which covers the substrate binding groove and needs to be proteolytically removed for protease activation[6].Some proteases carry a vacuolar targeting signal(NPIR) in the prodomain and a predicted endoplasmic reticulum protein retention signal(KDEL) at C-terminal region[7-8].Some PLCPs also carry a C-terminal granulin likedomain,which shares homology to granulins in animals,which are growth hormones released upon wounding[9].Plant PLCPs are phylogeneticallydivided into nine subfamilies based on phylogenetic analysis and conserved functional and structural features[1].
RD21(Responsive-to-Desiccation-21,AT1G47128) ofArabidopsisthalianabelongs to an intriguing class of PLCPs,which was initially found to be up-regulated indrought-stressed and typified by the presence of a C-terminal granulindomain[2,10-11].ArabidopsisRD21 is composed of fivedomains:an N-terminal signal peptide,an autoinhibitory prodomain,the proteasedomain,a proline-richdomain and a granulindomain[10].Localisation studies indicate that iRD21(immature RD21) is transported from the Endoplasmic Reticulum(ER) with ER bodies,small cellular organelles released from ER,in vacuoles,where conversion into mRD21(mature RD21) occurs[10,12].RD21-like proteases that carry a C-terminal granulindomain are found in manydifferent plant species including tomato[13],maize[14],potato[15]and radish[16].Transcript levels ofRD21 increases upondrought stress and high salt conditions,whiledoes not change upon treatments with heat,cold nor abscisic acid[2].Moreover,tomatoRD21-like gene,C14 andArabidopsisRD21 were highly expressed in leaf tissue and up-regulatedduring senescence[17].ThoughRD21 was extensively study inArabidopsis,a role ofRD21-like genes from no-model species is yet poorly understood.
In order todiscover the roles ofRD21 orthologous genes involving stress-responsive pathways in no-model species,we isolated and characterized aRD21 orthologous geneFeRD21 from buckwheat(F.esculentum),which is one of the oldestdomesticated crops of Asia,Europe and North America and is a multi-food-use pseudo-cereal with healing benefits[18].Moreover,we compared the RD21 and FeRD21 proteins structruedifference between buckwheat andArabidopsis.Furthermore,tissue specific expression ofFeRD21 gene from buckwheat was aslo analysed under various stresses.
1Materials and Methods
1.1 Plant material and stress treatments
Buckweat(F.esculentum,‘Xinong 9976’) seedlings were grown in 7 cm×7 cm×8 cm plastic pots filled with a commercial growing soil mix under a 16 h light/8 hdark photoperiod at 25 ℃ for 15days,and were subjected to various abiotic stresses.Seedlings were exposed to air on filter paper for induction ofdehydrationdrought response.To mimic salinity and ABA treatment,seedlings were transferred into solutions containing 200 mmol/L NaCl and 200 μmol/L ABA,respectively.Samples for above treatments were collected at 0,1,2,3,6,12,24 or 48 h after treatments.For senescent treat,seedlings were transferred to adark chamber with 75% humidity conditions at 25 ℃,samples were collected at 0,2,3,4,5,6,7 or 8d after treatment.After sampling atdifferent time points,seedlings weredropped immediately into liquid nitrogen and stored at -80 ℃ for RNA extraction,respectively.Leaves,stems and roots fromdifferent buckwheat plants grown under normal conditions were sampled,respectively.
1.2 Isolating FeRD21 from F. esculentum
Total RNA was extracted from seedlings using EASYspin Plus Kit according to the manufacturer’s protocol(Aidlab,China).First-strand cDNA was synthesized from 1 μg of thedNase I-treated RNA,using oligo(dT)15adaptor primer and M-MLV Reverse Transcriptase(TaKaRa,Japan).In order to isolate the RD21 homologous gene fromF.esculentum,a 486 bp fragment was amplified from the cDNA,prepared from leaves by using the forward primer FeRD21F(5′-TGTGGTAGTTGCTGGGCATTTTC-3′) and the reverse primer FeRD21R(5′-CCACGAGTTCTTCACAATCCAG-3′).Comparison with sequences in the NCBIdatabases revealed that the fragment was an internal coding region of an RD21 homologous gene.Isolation of the 3′ end ofFeRD21 was carried out using the 3′-full RACE Core Set Ver.2.0 kit(TaKaRa,Japan) following the protocol from the manufacturer with gene-specific primer GSPFeRD21(5′-GCCATTGACAGTGAAGA-TGATTAC-3′).The 5′ partial cDNA ofFeRD21 was isolated using the 5′ full-RACE Kit(TaKaRa,Japan) following the manufacturer’s protocol with the gene-specific primers FeRD21GSP1(5′-GTTAGCA-GTGCCAGCAAGGTTTC-3′) and FeRD21GSP2(5′-AACCATAACCAACAGCTGCAACAC-3′).The full-length cDNA ofFeRD21 was amplified with the primers of FeRD21F(5′-TCTCCACCACTGAAAAGCAG-AATC-3′) and FeRD21R(5′-AGGCGAATGAGTTGCACCATGAAC-3′).PCR was performed with a 5 min 94 ℃denaturation,followed by 30 cycles of 45 sdenaturating at 94 ℃,45 s annealing at 58 ℃,and 1 min extension at 72 ℃,with a final extension of 10 min.
1.3 Characterization of FeRD21
For phylogenetic assessment of the relationship of FeRD21(GenBank accession number:AFO83614) toArabidopsisproteases sequences,allArabidopsisPLCPs were obtained from TAIR and Genbank.Phylogenetic trees were constructed with MEGA 5.0 software using the Neighbor-Joining Method[19-20].Deduced amino acid sequences ofFeRD21 were also used for BLAST analysis on the GenBankdatabase.Based on Blast searches,multiple RD21A homologous proteases fromdifferent angiosperm lineages were selected for alignment.Full-length amino acid sequences containing N-terminal signal peptide,auto-inhibitory prodomain,proteasedomain,proline-richdomain and granulin-likedomain were aligned used the ClustalW program withdefault settings[10].The signal peptide of FeRD21 was predicted using SignalP 4.1 Server(http://www.cbs.dtu.dk/services/SignalP/).Moreover,the three essential residues(C,H,N) in the catalytic site triad was searched using the BLAST program(http://www.ncbi.nlm.nih.gov/BLAST/).
1.4 Expression analysis of FeRD21
For quantitative analysis,total RNA were isolated from treated seedlings asdescribed earlier.Quantitative real-time RT-PCR with three biological replicates was carried out with the gene specific primers QFeRD21F(5′-GATCATTGCTTAGGAAACACAATC-3′) and QFeRD21R(5′-CAGTAGATGTGAAGATGATAAATAGAG-3′) using an optical 96-well plate with an MJ research opticon TM2 system with SYBR green I and analyzed with the Bio-Rad CFX96 Optical System Software version 1.6.The reaction mixture was cycled as follows:95 ℃ for 3 min,followed by 40 cycles of 95 ℃ for 30 s,58 ℃ for 30 s,72 ℃ for 1 min.For the melt curve,we changed the temperature by increments of 0.5 ℃/s to 95 ℃.The experiments were repeated three times for each sample.TheF.esculentumactin was used as a positive control with the specific primers QFeactinF(5′-ACCTTGCTGGACGTGACCTTAC-3′) and QFeactinR(5′-CCATCAGGAAGCTCATAGTTC-3′).
2Results and analysis
2.1 Clone and classification of FeRD21 gene from F. esculentum
Full-length cDNA ofRD21 homologous gene fromF.esculentumwas obtained by homology-based cloning and RACE techniques following the above procedures.TheRD21 homologous gene fromF.esculentumtranscript was 1 750 bp and contained a 1 407 bp ORF(Open Reading Frame,ORF) encoding 468 amino acids,as well as a 87 bp 5′ untranslated region(5′-UTR) and a 256 bp 3′-UTR including a poly-A tail.The sequence wasdeposited in the GenBank under Accession Number JN605353.To comfirm the amplified fragment was the orthologous gene ofAtRD21 inF.esculentum,we performed a BLASTP search of thededuced RD21 homologous protein sequence to theArabidopsisprotein TAIR10database and found that RD21A(AT1G47128) was the closest orthologue.Furthermore,our phylogenetic analysis grouped the FeRD21 protein into PLCPs subfamily members whichcarry a C-terminal granulindomain(Fig.1)[1].Therefore,the gene is referred to asFeRD21(Fagopyrumesculentumresponsive todesiccation-21).
2.2 Structure of FeRD21
The protein alignmentdisplayed that FeRD21 has 468 amino acids(aa) containing a 25 aa N-terminal signal peptide(0-25),a 118 aa auto-inhibitory prodomain(26-143),a 211 aa proteasedomain(144-354),a 27 aa proline-richdomain(355-381) and a 59 aa granu-lin-likedomain(382-440)(Fig.2)[17,21-22].The prote-

Fig.1 Phylogenetic tree of Arabidopsis
asedomain of the FeRD21 was highly conserved,and harbored conservation of the catalytic residues Cys168-His304-Asn324within thisdomain[23].A GCNGG motif(206-210) was identified in FeRD21 and this motif is invariant in the cathepsin B-like proteinases[24].
2.3 Expression analysis of FeRD21
InF.esculentum,transcript levels ofFeRD21 in leaves and roots were obviously higher than stems of the seedlings grown at 25 ℃(Fig.3).Moreover,Quantitative real-time RT-PCR performeddisplayed that expression ofFeRD21 was up-regulated by salt,dehydration,ABA and senescent.With ABA treatment,FeRD21 expression became stronger within 48 h and the level of product peaked at 24 h(Fig.4,A).For se-nescent treatment,expression ofFeRD21 was increased

Fig.3 Expression of FeRD21 indifferent organs at 25 ℃

Fig.2 Sequence alignments of FeRD21 and others
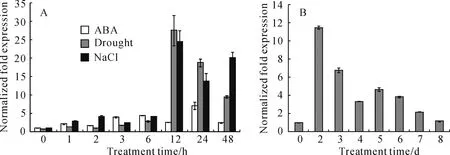
Fig.4 Expression of FeRD21 analyzed by qRT-PCR underdifferent abiotic stress conditions
rapidly,peaked at 2day,and thendecreased slowly from 3day to 8day(Fig.4,B).Underdehydration and NaCl treatment,FeRD21 transcript accumulated rapidly and reached its maximum at 12 h(Fig.4,A).
3Discussion
Cysteine protease enzymes belong to a large family of enzymes found in animals,plants,and microorganisms that play important roles in intracellular proteindegradation and organism PCD.Most plant cysteine proteases belong to the papain-like,metacaspase and legumain families.The papain-like cysteine proteases(PLCPs) are the most thoroughly investigated family among cysteine proteases[23].PLCPs are very stable enzymes and often found in proteolytically harsh environments such as the apoplast,the vacuole and lysosomes[1].Plant PLCPs can be classified into nine subfamilies based on conserved structural features:RD21A-like,CEP1-like,XCP2-like,XBCP3-like,THI1-like,SAG12-like,RD19A-like,AALP-like and CTB3-like.The C-terminal granulindomain occurs in two PLCP subfamilies:RD21A-like and XBCP3-like.
In this study,FeRD21 fromF.esculentum(buckwheat,Polygonaceae),which encoded a cysteine protease was isolated and characterized.Like other papain-like cysteine proteinases,three highly conserved catalytic residues of Cys168-His304-Asn324constituted the catalytic triad of cysteine proteases[24].Moreover,the C-terminal region of FeRD21 containing a granulindomain,which wasdifferent from other plant papain-like cysteine proteinases[1].So,the FeRD21 could be classificated into RD21A-like subfamilies.Moreover,A GCNGG motif was also identified in FeRD21 as invariant in the cathepsin B-like proteinases[25].
TheFeRD21 expressed throughout all plant vegetative organs,while transcript levels in leaves and roots were obviously higher than stems.InArabidopsis,RD21 genedisplayed similar expression patterns[1].Transcript levels ofRD21 in stems were obviously lower than leaves and roots.Moreover,the expression ofFeRD21 was up-regulated by many abiotic stresses,such as salt,dehydration,ABA,and senescent,which showed adifferent stress-responsive pathway withRD21 inArabidopsis.Although expression ofArabidopsisRD21 could be up-regulated bydrought stress and high salt conditions,transcript levels ofRD21do not change upon treatment with heat,cold nor abscisic acid.Moreover,theRD19 fromArabidopsisaslo showeddifferent stress responsive pathway withFeRD21.Previous study showd thatRD19 mRNAs were not induced by abscisic acid,cold and heat stress.On the other hand,transcription of theRD19 mRNAs was strongly induced under high-salt conditions,and theRD19 andRD21 were induced by changes in the osmotic potential of plant cells[2].In tomato,the transcript levels of aRD21 homogous geneC14 are induced by cold,drought andduring leaf senescence.Moreover,a 65 kD protein,matrue C14 protease,was found to be accumulated in the leaves ofdrought-stressed tomato(Lycopersiconesculentumcv.Starfire) plants.The protein level returns to control level when thedrought-stressed plants are rewatered.The protein was found to be mainly localized in the nuclei and chloroplasts ofdrought-stressed leaf cells[26].The RD21-homologue of potato,CYP,is transcriptionally induced in early stages ofPhytophthorainfestansinfection[27].Thedetail pathways ofFeRD21 involved in stress and immune responses should be furtherdiscovered inF.esculentum.
References:
[1]RICHAU K H,KASCHANI F,VERDOES M,etal.Subclassification and biochemical analysis of plant papain-like cysteine proteasesdisplays subfamily-specific characteristics[J].PlantPhysiol.,2012,158(4):1 583-1 599.
[2]KOIZUMI M,YAMAGUCHISHINOZAKI K,TSUJI H,etal.Structure and expression of two genes that encodedistinctdrought-inducible cysteine proteinases inArabidopsisthaliana[J].Gene,1993,129(2):175-182.
[3]BERNOUX M,TIMMERS T,JAUNEAU A,etal.RD19,anArabidopsiscysteine protease required for RRS1-R-mediated resistance,is relocalized to the nucleus by theRalstoniasolanacearumPopP2 effector[J].PlantCell,2008,20(8):2 252-2 264.
[4]DRENTH J,JANSONIUM J N,KOEKOEK R,etal.Structure of papain[J].Nature,1968,218(5 145):929-932.
[5]TURK V,TURK B,TURKd.Lysosomal cysteine protases:facts and opportunities[J].EMBOJ.,2001,20(17):4 629-4 633.
[6]TAYLOR M A,BAKER K C,BRIGGS G S,etal.Recombinant pro-regions from papain and papaya proteinase tv are selective high-affinity inhibitors of the mature papaya enzymes[J].ProteinEngineering,1995,8(1):59-62.
[7]GRUDKOWSKA M,ZAGDANSKA B.Multifunctional role of plant cysteine proteinases[J].ActaBiochimicaPolonica.,2004,51(3):609-624.
[8]ZHANG XM,WANG Y,LV X M,etal.NtCP56,a new cysteine protease inNicotianatabacumL.,involved in pollen graindevelopment[J].J.Exp.Bot.,2009,60(6):1 569-1 577.
[9]BATEMAN A,BENNETT H P.The granulin gene family:from cancer todementia[J].Bioessays,2009,31(11):1 245-1 254.
[10]YAMADA K,MATSUSHIMA R,NISHIMURA M,etal.A slow maturation of a cysteine protease with a granulindomain in the vacuoles of senescingArabidopsisleaves[J].PlantPhysiol.,2001,127(4):1 626-1 634.
[11]GU C,SHABAB M,STRASSER R,etal.Post-translational regulation and trafficking of the granulin-containing protease RD21 ofArabidopsisthaliana[J].PLoSOne,2012,7(3):e32422.
[12]CARTER C,PAN S,ZOUHAR J,etal.The vegetative vacuole proteome ofArabidopsisthalianareveals predicted and unexpected proteins[J].ThePlantCell,2004,16(12):3 285-3 303.
[13]DRAKE R,JOHN I,FARRELL A,etal.Isolation and analysis of cDNAs encoding tomato cysteine proteases expressedduring leaf senescence[J].PlantMolecularBiology,1996,30(4):755-767.
[14]YAMADA T,KONDO A,OHTA H,etal.Isolation of the protease component of maize cysteine protease-cystatin complex:release of cystatin is not crucial for the activation of the cysteine protease[J].PlantCellPhysiol.,2001,42(7):710-716.
[15]CHEN H J,HUANGd J,HOU W C,etal.Molecular cloning and characterization of a granulin-containing cysteine protease SPCP3 from sweet potato(Ipomoeabatatas) senescent leaves[J].JournalofPlantPhysiology,2006,163(8):863-876.
[16]KIKUCHI Y,SAIKA H,YUASA K,etal.Isolation and biochemical characterization of two forms of RD21 from cotyledons ofdaikon Radish(Raphanussativus)[J].JournalofBiochemistry,2008,144(6):789-798.
[17]SHINDO T,MISAS-VILLAMIL J C,HÖRGER A C,etal.A role in immunity forArabidopsiscysteine protease RD21,the ortholog of the tomato immune protease C14[J].PLoSOne,2012,7(1):e29317.
[18]CAWOY V,KINET J M,JACQUEMART A L.Morphology of nectaries and biology of nectar production in thedistylous speciesFagopyrumesculentum[J].Ann.Bot.,2008,102(5):675-684.
[19]TAMURA K,DUDLEY J,NEI M,etal.MEGA4:molecular evolutionary genetics analysis(MEGA) software version 4.0[J].Mol.Bio.Evol.,2007,24(8):1 596-1 599.
[20]TAMURA K,PETERSONd,PETERSON N,etal.MEGA5:molecular evolutionary genetics analysis using maximum likelihood,evolutionarydistance,and maximum parsimony methods[J].Mol.Biol.Evol.,2011,28(10):2 731-2 739.
[21]MARCHLER-BAUER A,LU S,ANDERSON JB,etal.CDD:a conserveddomaindatabase for the functional annotation of proteins[J].NucleicAcidsRes.,2011,39(1):D225-229.
[22]PETERSEN TN,BRUNAK S,VON HEIJNE G,etal.SignalP 4.0:discriminating signal peptides from transmembrane regions[J].Nat.Methods.,2011,8(10):785-786.
[23]ZHANGd,LIUd,LV X,etal.The cysteine protease CEP1,a key executor involved in tapetal programmed celldeath,regulates pollendevelopment inArabidopsis[J].PlantCell,2014,26(7):2 939-2 961.
[24]MARCHLER-BAUER A,ZHENG C,CHITSAZ F,etal.CDD:conserveddomains and protein three-dimensional structure[J].NucleicAcidsRes.,2013,41(1):D348-352.
[25]BEYENE G,FOYER C H,KUNERT K J.Two new cysteine proteinases with specific expression patterns in mature and senescent tobacco(NicotianatabacumL.) leaves[J].JournalofExperimentalBotany,2006,57(6):1 431-1 443.
[26]HARRAK H,AZELMAT S,BAKER EN,etal.Isolation and characterization of a gene encoding adrought-induced cysteine protease in tomato(Lycopersiconesculentum)[J].Genome,2001,44(3):368-374.
[27]AVROVA A O,STEWART H E,DE JONG W,etal.A cysteine protease gene is expressed early in resistant potato interactions withPhytophthorainfestans[J].MolecularPlant-MicrobeInteractions,1999,12(12):1 114-1 119.
(编辑:宋亚珍)

