Changes of TIZ expression in epithelial ovarian cancer cells
Huan-Yu Zheng, Hong-Yu Zheng, Yun-Tao Zhou, En-Ling Liu*, Jie Li, Yan-Mei Zhang
1Department of Obstetrics & Gynecology, Affiliated Tangshan Workers Hospital of Hebei Medical University, Tangshan, Hebei 063000, China
2Department of Pharmacy, Tangshan People's Hospital, Tangshan, Hebei 063000, China
3Central Laboratory, Affiliated Tangshan Workers Hospital of Hebei Medical University, Tangshan, Hebei 063000, China
Changes of TIZ expression in epithelial ovarian cancer cells
Huan-Yu Zheng1, Hong-Yu Zheng2, Yun-Tao Zhou3, En-Ling Liu1*, Jie Li1, Yan-Mei Zhang1
1Department of Obstetrics & Gynecology, Affiliated Tangshan Workers Hospital of Hebei Medical University, Tangshan, Hebei 063000, China
2Department of Pharmacy, Tangshan People's Hospital, Tangshan, Hebei 063000, China
3Central Laboratory, Affiliated Tangshan Workers Hospital of Hebei Medical University, Tangshan, Hebei 063000, China
ARTICLE INFO
Article history:
Received 15 November 2014
Received in revised form 20 December 2014
Accepted 15 January 2015
Available online 20 February 2015
TIZ expression
Epithelial ovarian cancer cells
Cell proliferation
Invasion ability
Objective: To study the change of TIZ expression in epithelial ovarian cancer cells. Methods: HO8910 cells were transinfected with siRNA to inhibit the expression of TIZ. pcDNA3.1-TIZ vectors were combined to increase the TIZ expression level. The cell viability, colony forming efficiency and cycle distribution of HO8910, HO8910/NC, HO8910/pcDNA3.1-NC, HO8910/ TIZ-573 and HO8910/pcDNA3.1-TIZ were compared, and the invasion rate, migration rate and adhesion rate between 5 groups of cells were compared. Results: Compared with those of HO8910, HO8910/ NC and HO8910/pcDNA3.1-NC, the cell viability, colony forming efficiency and cell cycle distribution of HO8910/ TIZ-573 were increased, while the indexes of HO8910/pcDNA3.1-NC were decreased with statistical significant difference (P<0.05). There was no statistical significant difference in the invasion rate, migration rate and adhesion rate between 5 groups of cells (P>0.05). Conclusions: The expression of TIZ can inhibit the proliferation of epithelial ovarian cancer cells.
1. Introduction
The epithelial ovarian tumor is a common malignant gynecological tumor, occupying 50%-70% of ovarian tumors [1]. With the advances in the modern cellular and molecular biology, we have gained a deeper understanding in the occurrence and development of the epithelial ovarian cancer. Considerable research has indicated that such a tumor is the result of a combination of many genes and factors in different stages[2]. TRAF-6 inhibitory zinc finger protein (TIZ) belongs to the C2H2-type zinc finger protein family, which can be combined with the tumor necrosis factor receptor-associated factor 6 (TRAF-6) to play a role in the regulatio[3,4]. According to clinical research, the expression level of TIZ protein in the serum of patients with ovarian malignant tumor was significantly higher than the one of patients with ovarian benign tumor and the one of normal subjects, which indicates the high correlation between the expression level of TIZ and the ovarian cancer[5]. In this study, we observed the expression level of TIZ gene in epithelial ovarian tumor cells (HO8910) to study the effect of TIZ on the biological characteristics of tumor cells.
2. Materials and methods
2.1. Materials and reagents
The epithelial ovarian tumor cells (HO8910) provided by Shanghai SXBio Biotechnology Co., Ltd. was chosen as the experimental subject. pGPU6/GFP/Neo interference vector was provided by Shanghai GenePharma Co., Ltd. pcDNA3.1 expression vector, TRIzol, propidium iodide (PI), RPMI 1640 medium and liposome LipofectAMINETM2000 were all purchased from Invitrogen, a brand under the Life Technologies brand of the Thermo Fisher Scientific corporation. cDNA synthesis kit was provided by MBI Fermentas; DNA segment rapid purification recycle kit was provided by Beijing Sunbiotech Co., Ltd.; E.Z.N.A.® Plasmid Mini Kit I was provided by Omega Biotek. The fibronectin (FN), transwell chamber and artificial basement membrane matrigel were all provided by Coming Costar. Escherichia coli (E. coli) DH-5α was preserved in this laboratory.
2.2. Determination of TIZ expression level in HO8910 cell lines
TRIzol reagent was used to extract mRNA of HO8910 cells for the reverse transcription. cDNA amplification was performed with the reaction system of 25 μL. The reaction conditions were the degeneration at 94 ℃ for 5 min, at 94 ℃ for 1 min, at 5 ℃ for 1 min and at 72 ℃ for 1 min, 35 times of sequence cycles, extension at 72 ℃ for 5 min and the preservation of product at 4 ℃. The electrophoretic determination of PCR products was performed using 1% agarose gel. All the above primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd.
2.3. Interference of small RNA in TIZ expression
Three segments of small interfering RNA (siRNA) were designed and synthesized according to the mRNA sequences of TIZ in GenBank: the upstream segment of TIZ-554 was 5'-CACCUAACUCGACAUGAAATT-3' and the downstream one was 5'-UUUCAUGUCGAGUUAGGUGTG-3'; the upstream segment of TIZ-573 was 5'-GAAAUUAUACCAAGGUGAATT-3' and the downstream one was 5'-UUCACCUUGGUAUAAUUUCTT-3'; the upstream segment of TIZ-620 was 5'-GCUGUUAACCAAUCUUCAATT-3' and the downstream one was 5'-UUGAAGAUIJCCImAACAGCTT-3'. Negative control (NC): the upstream segment was 5'-UUCUCCGAACGUCUCACGUTT-3' and the downstream one was 5'-ACGUGACACGUUCGGAGAATT-3'. All the above sequences were synthesized by Shanghai GenePharma Co., Ltd.
TIZ-554, TIZ-573, TIZ-620 and negative control sequences were combined with T4-DNA ligase and pGPU6/GFP/Neo interference vectors respectively. E. coli DH-5α competent cells were transformed by products respectively. The bacterial flora was screened using 100 μg/uL ampicillin and it was cultured for amplification. The plasmid was extracted and PCR and double restriction enzyme digestion were performed again. The positive recombinant plasmid was used for bi-directional sequencing of DNA.
HO8910 cells were transfected using the recombinant plasmids of TIZ-554, TIZ-573, TIZ-620 and NC, named as HO8910/TIZ-554, HO8910/TIZ-573, HO8910/TIZ-620 and HO8910/NC respectively. The above cells and untreated HO8910 cells were compared for the expression level of TIZ.
2.4. Upregulation of TIZ expression via in vitro transfection
PCR product and pcDNA3.1 vector were both treated by double digestion of BamH Ⅰ and EcoR Ⅰ and the products of digestion were separated. Target fragments were recovered and purified. And the recombinant plasmid was constructed. T4-DNA ligase was used to combine the vector and the target gene. E. coli DH-5α competent cells were transformed by the product (pcDNA3.1-TIZ). The extraction and detection of recombinant plasmid were the same as above. HO8910 cells were transfected using pcDNA3.1-TIZ plasmid and pcDNA3.1 plasmid, named as HO8910/pcDNA3.1-TIZ cell and HO8910/pcDNA3.1-NC cell respectively. mRNA of untreated HO8910 cells, HO8910/ pcDNA3.1-TIZ cells and HO8910/pcDNA3.1-NC cells was extracted respectively and RT-PCR was used to detect the expression level of TIZ in cells.
2.5. Detection of cell viability by MTT method
Cells in the logarithmic phase were taken to prepare the single cell suspension, with a cell density of 1×105/mL. A total of 100 μL of cell suspension were added in the 24-well plate, with 3 repeated wells. The control group was set with 100 μL of medium in each well. A total of 100 μL of 5 mg/ mL MTT solution was added in each well and incubated at 37 ℃ for 4 h. The liquid was removed, 500 μL DMSO was added in each well and oscillated for 10 min. Absorbance (A) value was measured at 450 nm and the growth curve of cells was drawn.
2.6. Colony formation assay
Stably transfected cells were chosen to prepare the single cell suspension. Cells were seeded in the 24-well plate, with 100 cells in each well. The 24-well plate was gently shaken to achieve the uniform distribution of cells. Cells were put in the thermotank of 37℃ with 5% CO2volume fraction for the culture of 5-7 days. After the culture, cells were washed using PBS. Cells were stained by Giemsa. The number of colony forming units was counted under the optical microscope. A total of 15-50 cells were taken as a colony. The equation was as follows: Colony-forming efficiency (%) = Number of clones / Number of inoculated cells × 100%.
2.7. Determination of changes in cell cycle using flow cytometry
Cells were taken in the logarithmic phase to prepare the single cell suspension. A total of 15 mL of 95% ethanol solution pre-cooled at -20 ℃ was added and fixed at 4 ℃ for 1 h. The ethanol solution was removed using the centrifugal method. The cell density was adjusted to 1×106/mL, digested using 50 μg/mL RNA enzyme for 2 min and strained by using 50 μg/mL PI away from light for 30 min. The content distribution of DNA was analyzed by using flow cytometry, as well as the proportion of cells in phases of G0/ G1, G2+M and S. The proportion of cells in the phase of G0/G1was taken as the indicator to reflect the proliferation ability of cells.
2.8. Changes in in vitro invasion and migration
Polycarbonate membrane filter of Transwell was treated using PBS with 5 μg FN. A total of 50μL of 1.25mg/ mL matrigel was added in the upper cavity. The treated
Transwell was incubated at 37 ℃ for 4-5 h. The cell suspension was added in the upper and lower cavity. After 24 h, MTT method was used to detect D value of adhesion cells in upper and lower cavities at 450 nm. The equation was as follows:Invasion rate (%) = D450nmof Cells in lower cavity/D450nmof cells in upper cavity × 100% [6].
Except the addition of matrigel, the determination of in vitro migration was similar with the in vitro invasion assay. Migration rate (%) = D450nmof cells in lower cavity/D450nmof cells in upper cavity × 100%[6].
2.9. Changes in in vitro adhesion ability
The 96-well plate was taken, and 50 μL of 20 mg/L FN was added in each well, allowing it to air dry on the clean bench over the night, and then the plate was put in the refrigerator at 4 ℃. Before using, it was used by PBS twice. The binding site was closed, the cell suspension was added, cultured for 1 h and washed by PBS to remove the unadhered cells. MTT method was used to detect D value of adhesion cells in upper and lower cavities at 450 nm. Adhesion rate (%) = D450nmof adhesion cells in experiment group/D450nmof adhesion cells in control group × 100%[6]. Repeat the procedures (2.4-2.8) for three times.
2.10. Statistical analysis
Statistical analysis was performed by using SPSS15.0. The measurement data were expressed as mean±sd. The oneway ANOVA was used to analyze the difference between groups, while the analysis of variance with repeated measures was used in comparison of cell viability at different times, P<0.05 indicating the statistical significant difference.
3. Results
3.1. Determination of siRNA interference efficiency
Compared with NC sequence transfected cells, the expression level of TIZ in three siRNA transfected fragments of TIZ-554, TIZ-573 and TIZ-620 was significantly decreased (P<0.05); the expression level of TIZ in TIZ-573 transfected cells was significantly lower than others (P<0.05). There was no statistical difference in the expression level of TIZ between HO8910/NC and HO8910 (P>0.05), as shown in Figure 1.
3.2. TIZ expression in pcDNA3.1-TIZ transfected cells
There was no statistical difference in the TIZ expression between HO8910 and HO8910/pcDNA3.1-NC (P>0.05), but TIZ expression in HO8910/pcDNA3.1-TIZ cells was significantly increased (P<0.05), as shown in Figure 2.
3.3. Effect of TIZ expression on cell viability
TIZ-573siRNA fragments with the best interference effect were chosen for the study. There was no statistical difference in the cell viability between HO8910, HO8910/ NC and HO8910/pcDNA3.1-NC (P>0.05). The cell viability of HO8910/ TIZ-573 was significantky increased, while the one of HO8910/pcDNA3.1-TIZ was significantly decreased (P<0.05), as shown in Figure 3.
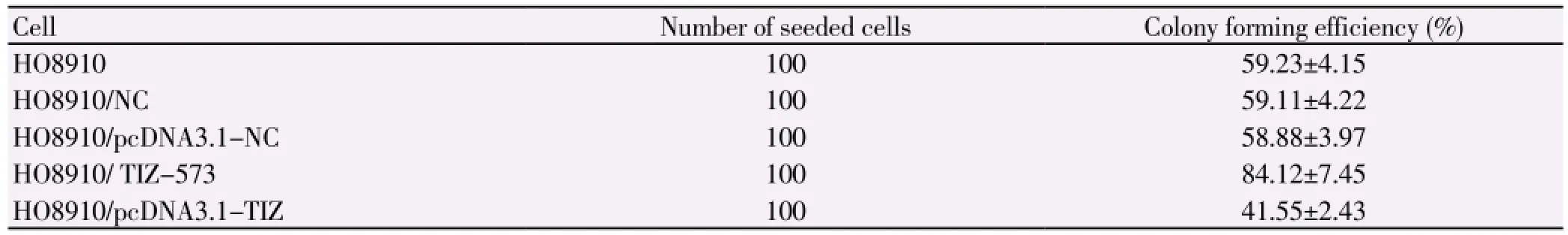
Table 1 Colony forming efficiency of different cells (mean±sd).
3.4. Effect of TIZ expression on colony forming efficiency
There was no statistical difference in the colony forming efficiency between HO8910, HO8910/NC and HO8910/
pcDNA3.1-NC (P>0.05). The colony forming efficiency of HO8910/ TIZ-573 cell was increased, while the one of HO8910/pcDNA3.1-TIZ cell was decreased, with the statistical difference (P<0.05), as shown in Table 1.
3.5. Effect of TIZ expression on cell cycle
There was no statistical difference in the percentage of cells in different phases between HO8910, HO8910/NC and HO8910/pcDNA3.1-NC (P>0.05). Cells of HO8910/ TIZ-573 in the phase of G0/G1were significantly increased, while cells of HO8910/pcDNA3.1-TIZ in the phase of G0/G1were significantly decreased (P<0.05), as shown in Table 2.
3.6. Effect of TIZ expression on in vitro invasion, migration and adhesion of cells
There was no statistical difference in the in vitro invasion rate, migration rate and adhesion rate between HO8910, HO8910/NC, HO8910/pcDNA3.1-NC, HO8910/ TIZ-573 and HO8910/pcDNA3.1-TIZ (P>0.05), as shown in Table 3.
4. Discussion
According to the clinical studies, the expression level of TIZ protein in the serum of patients with ovarian malignant tumor was significantly higher than the one of patients with ovarian benign tumor and the one of normal subjects, which indicates the high correlation between the expression level of TIZ and the ovarian cancer. In this study, we used siRNA to interfere with the expression level of TIZ in HO8910 and then constructed the expression vector to increase the expression level of TIZ. Relying on the in vitro experiment, we studied the effect of change in the expression level of TIZ on the biological characteristics of epithelial ovarian tumor cells (HO8910).
In the study, compared with that in transfected cells of NC sequence, the TIZ expression in three segments of cells transfected by siRNA, namely TIZ-554, TIZ-573 and TIZ-620, was significantly decreased, while the transfection efficiency of TIZ-573 was the highest; the TIZ expression in HO8910/pcDNA3.1-TIZ cell was significantly increased, which indicates the success of interference in the TIZ expression in HO8910.

Table 2 Cell cycle of different cells (mean±sd, %).

Table 3 In vitro invasion, migration and adhesion of different cells (mean±sd, %).
TIZ-573siRNA segment with the best interference effect was selected for the further study. The cell viability, colony forming efficiency and cell cycle of HO8910, HO8910/NC, HO8910/pcDNA3.1-NC, HO8910/ TIZ-573 and HO8910/ pcDNA3.1-TIZ were compared and then the proliferation ability of different cells was evaluated. Compared with that of HO8910, HO8910/NC and HO8910/pcDNA3.1-NC, the cell viability of HO8910/ TIZ-573 was increased, as well as the colony forming efficiency and the number of cells in the phase of G0/G1; while the cell viability of HO8910/pcDNA3.1-TIZ was decreased, as well as the colony forming efficiency and the number of cells in the phase of G0/G1. It indicates that the expression of TIZ inhibits the proliferation of epithelial ovarian cancer cells. TIZ is just like the hospital of zinc finger protein KRAB family. Such kind of protein all plays a critical role in the embryonic development, transformation and differentiation of cells and the regulation of cell cycles[7,8]. The mechanism for TIZ to inhibit the proliferation of tumor cells has not been clear, but some research has indicated that it can effectively inhibit the TRAF-6 mediated signal transduction. It combines with N terminal of TRAF-6 to change the protein conformation of TRAF-6 and then blocks the transduction of downstream signals[9]. The TRAF-6 mediated signal transduction is
relatively complicated, mainly including two ways[10]: (1) TRAF-6→mitogen-activated protein kinase kinase (MAPKK)→mitogen-activated protein kinase (MAPK); (2)TRAF-6→MAPKK→nuclear factor KB (NF-κB). The downstream signal proteins involved in the above two ways, such as JNK, P38, ERK1/2, C-FOS and NF-κB, are all closely related to the occurrence and development of ovarian cancer[11,12]. By blocking the TRAF-6 mediated signal transduction, TIZ can significantly inhibit the expression of the above proteins and thus inhibit the proliferation of epithelial ovarian cancer cells (HO8910).
In this study, there was no statistical difference in the invasion rate, migration rate and adhesion rate between HO8910, HO8910/NC, HO8910/pcDNA3.1-NC, HO8910/ TIZ-573 and HO8910/pcDNA3.1-TIZ, which indicates that there is no significant effect of TIZ expression on the in vitro invasion, migration and adhesion of epithelial ovarian cancer cells. The invasion ability of ovarian malignant tumor was relatively strong. 76% diagnosed patients had a wide migration of the abdominal cavity, the pelvic plant and the liver substance or the pleura and the brain[13]. The migration is the result of combination of cell adhesion and new blood vessels. Presently, there has been no definite conclusion on the correlation between TIZ expression and the spread and migration of epithelial ovarian tumor. Some research indicated that in the TIZ-mediated signal transduction, there was a high correlation between the cell factors of lysophosphatidic acid, matrix metalloproteinases, urokinase-type plasminogen activator, vascular endothelial growth factor and intercellular adhesion molecule and the invasion and migration of ovarian cancer[14,15]. The downstream signal molecules of TIZ expression had a negative feedback to the expression of TIZ, which then decreased the expression level of TIZ and weakened the inhibition ability; the decrease in the expression level of TIZ would also inhibit the activation of downstream signaling pathways to deactivate the signal molecules related to the invasion and migration of tumor and then decrease its invasion ability[16,17]. Anyway, TIZ possesses the dualdirection regulation on the invasion ability of HO8910. In conclusion, this study indicates that TIZ expression can inhibit the proliferation of epithelial ovarian tumor, but has no obvious effect on the invasion ability of tumor.
Conflict of interests
We declare that we have no conflict of interest.
[1] Pan XL.Expression of k-ras point mutation and p53 protein in ovarian carcinoma. Chin J Clin Exp Path 2005; 21(1): 66-69, 72.
[2] Fujita M, Enomoto T, Murata Y. Genetic alterations in ovarian carcinoma: with specific reference to histological subtypes. Mol Cell Endocrinol 2003; 202(1-2): 97-99.
[3] Zhao BB, Zhang W, Wang Q, Yang ZJ, Li L. The effect of TIZ gene overexpression on biological characteristics of epithelial ovarian cancer cells .Tumor 2012; 32(1): 21-26.
[4] Kar S, Ukil A, Das PK. Cystatin cures visceral leishmaniasis by NF-kappaB-mediated proinflammatory response through co-ordination of TLR/MyD88 signaling with p105-Tpl2-ERK pathway. Eur J Immunol 2011; 41(1): 116-127.
[5] Muffak-Granero K, Olmedo C, Garcia-Alcalde F, Comino A, Villegas T, Villar JM, et al. Gene network profiling before and after transplantation in alcoholic cirrhosis liver transplant recipients. Transplant Proc 2012; 44(6): 1493-1495.
[6] Nakayama N, Nakayama K, Yeasmin SK.RAS or BRAF mutation status is a useful predictor of sensitivity to MEK inhibition in ovarian cancer. Br J Cancer 2008; 99(12): 2020-2028.
[7] Zhao BB, Zhang W, Wang Q,Yang ZJ, Li L. Biological effects of down-regulating of TIZ gene on ovarian cancer cells in vitro. Chin J Obs Gyn 2012; 47 (10): 781-786.
[8] Zhao BB, Zhang W, Wang Q, Li L, Yang ZJ. Full-length amplification of TIZ gene and construction of eukaryotic expressing vector with such gene. J Guangxi Med Uni 2012; 29(1): 45-47.
[9] Avila M, Martinez-Juarez A, Ibarra-Sanchez A, Gonzalez-Espinosa C. Lyn kinase controls TLR4-dependent IKK and MAPK activation modulating the activity of TRAF-6/TAK-1 protein complex in mast cells. Innate Immun 2012; 18(4): 648-660.
[10] Walsh DE, Greene CM, Carroll TP, Taggart CC, Gallagher PM, O'Neill SJ, et al. Interleukin-8 up-regulation by neutrophil elastase is mediated by MyD88/IRAK/TRAF-6 in human bronchial epithelium. J Biol Chem 2001; 276(38): 35494-35499.
[11] Jefferies C, Bowie A, Brady G, Cooke EL, Li X, O'Neill LA. Transactivation by the p65 subunit of NF-kappaB in response to interleukin-1 (IL-1) involves MyD88, IL-1 receptor-associated kinase 1, TRAF-6, and Rac1. Mol Cell Biol 2001; 21(14): 4544-4552.
[12] Schwandner R,Yamaguchi K, Cao Z. Requirement of tumor necrosis factor receptor-associated factor (TRAF)6 in interleukin 17 signal transduction. J Exp Med 2000; 191(7): 1233-1240.
[13] Liu CZ, Zhang L, Chang XH, Cheng YX, Cheng HY, Ye X, et al. Overexpression and immunosuppressive functions of transforming growth factor 1, vascular endothelial growth factor and interleukin-10 in epithelial ovarian cancer. Chin J Cancer Res 2012; 24(2): 130-137.
[14] Zhang G, Li XP, Liu BJ, Wang JL, Wang SJ, Cui H, et al. Oxaliplatin-based combination chemotherapy is still effective for the treatment of recurrent and platinum-resistant epithelial ovarian cancer: results from a single center. Chin Med J (Engl) 2013; 126(23): 4477-4482.
[15] Jiang X, Zhu T, Yang J, Li S, Ye S, Liao S, et al. Identification of novel epithelial oOvarian cancer biomarkers by cross-laboratory microarray analysis. J Huazhong Univ Sci Technol Med Sci 2010; 30(3): 354-359.
[16] Yu L, Wu SW, Song WQ, Zhou L, Cheng ZN. Vasculogenic mimicry and expression of E-cadherin in epithelial ovarian cancer and its clinical significance. Chin J Path 2011; 27(11): 2120-2125.
[17] Xiao J, Li JD, Xu MM, Zhu AN, Feng YL. Clinical characteristics of umbilical migration of epithelial ovarian cancer. National Med J Chi 2013; 93(25): 1986-1988.
ment heading
10.1016/S1995-7645(14)60308-4
*Corresponding author: En-Ling Liu, PhD, Chief Physician, Affiliated Tangshan Workers Hospital of Hebei Medical University, Tangshan, Hebei 063000, China.
Tel: 13832828669
E-mail: luienlin669@126.com
Foundation project: It is supported by Soft Science Research Project in Hebei Province (12457612).
 Asian Pacific Journal of Tropical Medicine2015年2期
Asian Pacific Journal of Tropical Medicine2015年2期
- Asian Pacific Journal of Tropical Medicine的其它文章
- Effect of interferon plus ribavirin therapy on hepatitis C virus genotype 3 patients from Pakistan: Treatment response, side effects and future prospective
- Imported cases of dengue fever in Russia during 2010-2013
- Detection and characterization of Chlamydophila psittaci in asymptomatic feral pigeons (Columba livia domestica) in central Thailand
- Chemical composition of Rosmarinus and Lavandula essential oils and their insecticidal effects on Orgyia trigotephras (Lepidoptera, Lymantriidae)
- Total phenolic content, in vitro antioxidant activity and chemical composition of plant extracts from semiarid Mexican region
- Cytoprotective and anti-inflammatory effects of kernel extract from Adenanthera pavonina on lipopolysaccharide-stimulated rat peritoneal macrophages
