Antibiotic susceptibility profiling and virulence potential of Campylobacter jejuni isolates from different sources in Pakistan
Fariha Masood Siddiqui, Muhammad Akram, Nighat Noureen, Zobia Noreen, Habib Bokhari
Department of Biosciences, COMSATS Institute of Information Technology, Park Road, 44000, Islamabad, Pakistan
Antibiotic susceptibility profiling and virulence potential of Campylobacter jejuni isolates from different sources in Pakistan
Fariha Masood Siddiqui, Muhammad Akram, Nighat Noureen, Zobia Noreen, Habib Bokhari*
Department of Biosciences, COMSATS Institute of Information Technology, Park Road, 44000, Islamabad, Pakistan
ARTICLE INFO
Article history:
Received 24 December 2014
Received in revised form 10 January 2015
Accepted 15 February 2015
Available online 20 March 2015
Campylobacter jejuni
Antibiotic susceptibility
Virulence genes
PCR
RAPD
Objective: To determine antibiotic resistance patterns and virulence potential of Campylobacter jejuni (C. jejuni) isolates from clinical human diarrheal infections, cattle and healthy broilers. Methods: Antibiotic sensitivity patterns of C. jejuni isolates were determined by Kirby Bauer Disc Diffusion assay. These isolates were then subjected to virulence profiling for the detection of mapA (membrane-associated protein), cadF (fibronectin binding protein), wlaN (beta-1,3-galactosyltransferase) and neuAB (sialic acid biosynthesis gene). Further C. jejuni isolates were grouped by random amplification of polymorphic DNA (RAPD) profiling. Results: A total of 436 samples from poultry (n=88), cattle (n=216) and humans (n=132) from different locations were collected. Results revealed percentage of C. jejuni isolates were 35.2% (31/88), 25.0% (54/216) and 11.3% (15/132) among poultry, cattle and clinical human samples respectively. Antibiotic susceptibility results showed that similar resistance patterns to cephalothin was ie. 87.0%, 87.1% and 89%among humans, poultry and cattle respectively, followed by sulfamethoxazole+trimethoprim 40.0%, 38.7% and 31.0% in humans, poultry and cattle and Ampicillin 40%, 32% and 20% in humans, poultry and cattle respectively. Beta-lactamase activity was detected in 40.00% humans, 20.37% cattle and 32.25% in poultry C. jejuni isolates. CadF and mapA were present in all poultry, cattle and human C. jejuni isolates, wlaN was not detected in any isolate and neuAB was found in 9/31 (36%) poultry isolates. RAPD profiling results suggested high diversity of C. jejuni isolates. Conclusions: Detection of multidrug resistant C. jejuni strains from poultry and cattle is alarming as they can be potential hazard to humans. Moreover, predominant association of virulence factors, cadF and mapA (100 % each) in C. jejuni isolates from all sources and neuAB (36%) with poultry isolates suggest the potential source of transmission of diverse types of C. jejuni to humans.
1. Introduction
Campylobacter jejuni (C. jejuni) is an important food-born zoonotic pathogen, and one of the leading causes of human food borne illnesses (Campylobacteriosis) worldwide[1,2]. The most important source of transmission of this pathogen to humans is through contaminated animal products, especially poultry meat as well as direct contact with cattle shedding C. jejuni, or handling raw or undercooked poultry[3,4]. Campylobacter has been reported from broiler flocks in various European countries at the prevalence rates ranging from 38.1% to 79.2%[5,6]. Antibiotics play a vital role in human and veterinary medicine for treatment and prevention of infections but are also used as growth promoters in food animals[7]. Their increased use has resulted in the increased incidences of infection with enteric bacteria with higher levels of antibiotic resistance[8]. Campylobacter spp. has developed resistance to many clinically important antimicrobials, including fluoroquinolones (FQ) during the recent past[9-12]. It is believed that their transmission and spread are not only affected by the environmental and host factors, but also are influenced by the relative fitness of the drug-resistantorganisms in the absence of selection pressure[13]. Campylobacter spp. has shown resistance to a large number of beta lactam antimicrobial agents. However, the behaviours of others, such as ampicillin and some of the expanded-spectrum cephalosporins, are variable and not very clearly defined[14].
The current study gives the perspective of distribution of multiple antibiotic resistance, beta lactamase activity and virulence attribution to C. jejuni isolates from clinical human diarrheal infections, cattle and healthy broilers sharing the environment with the humans from the heavily populated city of Rawalpindi, Pakistan and its suburbs. Furthermore, their random amplification of polymorphic DNA (RAPD) profiling was carried out for determining their diversity.
2. Materials and methods
2.1. Sampling
This study was carried out from December 2011 and December 2012. The samples were collected from poultry slaughter houses from one of the country's leading poultry producer city, cattle farms and human clinical diarrheal cases. A total of 436 samples collected consisting of 216 cattle faecal samples, 132 human clinical samples and 88 poultry samples. Samples were collected in sterile cotton swabs containing Carry-Blair medium and transported to Microbiology Laboratory of COMSATS Institute of Information Technology, Islamabad.
2.2. Culturing and isolation
The samples collected were streaked onto modified Charcoal Cefoperazone Deoxycholate Agar (mCCDA) (Oxoid, CM0739) containing CCDA selective supplement (Oxoid, SR0155). Samples were incubated in 2.5 litres airtight jar along with Campygen sachets (Oxoid, CN025A) to generate microaerophilic condition at 42 ℃ for 48-72 hours. Suspected Campylobacter colonies were subcultured on Muller Hinton Agar (Oxoid, CM0337) with addition of 5% sheep blood[15].
2.3. Biochemical identification
Positive growth of Campylobacter isolates was further subjected to standard biochemical tests consisting of oxidase, catalase, indoxyl acetate and hippurate. In the case of indoxyl-acetate test, change of colour from colourless to blue green indicative of the presence of Campylobacter spp. and in case of hippurate hydrolysis, development of blue/purple colour in hippurate solution indicated positive reaction for presence of C. jejuni with the production of hippuricase enzyme and clear or grey colouration indicate negative reaction for its presence. A positive test for both reactions was indicative of C. jejuni[16].
2.4. Molecular detection of C. jejuni
Bacterial DNA was obtained by whole-cell lysate method as described by Singh et al. Primers used for confirmation of C. jejuni by PCR were MDS-16S rRNA (targeting 16S RNA gene), hipO (Hippurate hydrolysis gene) as described in Table 1. PCR was performed as previously described[17,18]. Amplified PCR products were analyzed on 1.5% agarose gel stained with ethedium bromide.
2.5. Virulence typing
C. jejuni isolates were screened for the presence of virulence genes (Table 1). Primers were designed against C. jejuni adhesin, cadF (fibronectin binding protein) (400 bp) gene, wlaN (putative beta-1,3-galactosyltransferase) (330 bp), neuAB (sialic acid biosynthesis gene) (755 bps) and mapA (membraneassociated protein) (94 bps).
2.6. RAPD PCR
For RAPD analysis of C. jejuni OPA11 primer was used as described by Hernandez et al. Briefly, the reaction mixture was carried out in a total volume of 25 μL containing 40 ng total DNA of each strain, 1.36 pM primer), 1.6 U Taq DNA polymerase (Super Taq), 1.5 μL 500 mM MgCl2, 0.7 μL 10 mM dNTPs in 1X PCR buffer (Fermentas). The PCR products were then separated by 1.5% agarose gel electrophoresis and visualized by ethidium bromide staining. Dendrogram was constructed using DendroUPGMA (genomes.urv.cat/UPGMA/) for RAPD PCR analysis.
2.7. Antimicrobial susceptibility profiling and beta-lactamase detection
Antibiotic susceptibility profiling was carried out using chloramphenicol C (30 μg), tetracycline (TE) (30 μg), streptomycin (S) (10 μg), ciprofloxacin (CIP) (5 μg), amoxicillin clavulenic (AMC) acid (30 μg), nalidixic acid (NA) (30 μ g), erythromycin (E) (30 μg), gentamycin (CN) (10 μg), sulphomethoxazole + trimethoprim (SXT) (25 μg) and cephalothin (CEF), respectively (Oxoid, UK) as described by Gaudreau et al[19]. Analysis of zone diameter was done according to the CLSI (2010). Beta-lactamases were detected by use of Cefinase disks (BBL Microbiology Systems) as described by Lachance et al[20].
3. Results
3.1. PCR confirmation of Campylobacter isolates
Biochemically verified C. jejuni strains were further subjected to PCR using primers against conserved 16S rRNA (amplification product 857 bps) and hip gene (hippurate hydrolysis gene) (344 bps).

Table 1 Primers for identification and virulotyping of C. jejuni.
3.2. C. jejuni distribution pattern among various sources
Four hundred and thirty-six samples were analysed in this study. 100 out of a total of 436 samples were confirmed as C. jejuni ie., the overall prevalence rate was 100/436 (22.93%). The isolation rate of C. jejuni was (n=31) 35.2%, (n=54) 25.0% and (n=15) 11.3% in poultry, cattle and humans, respectively.
3.3. Antimicrobial susceptibility profile
Antibiotic resistance profile of C. jejuni isolates from humans, cattle and poultry sources was determined using 10 antibiotics according to CLSI 2010. Comparison of antibiotic susceptibilities of C. jejuni isolates from different sources is shown in Table 2. Antibiotic susceptibility of the isolates revealed that resistance to cephalothin was the most common ie. 87.0%, 89.0% and 87.1%, followed by trimethoprim/sulfamethoxazole 40.0%, 38.7% and 31.0% and amoxicillin clavulenic acid 40%, 32% and 20% in human, cattle and poultry respectively (Figure 1). Multidrug resistance was also identified in strains from different sources (Table 3). Gentamicin was found to be the most sensitive antibiotic with resistance of 7%, 0% and 9% in humans, poultry and cattle isolates.
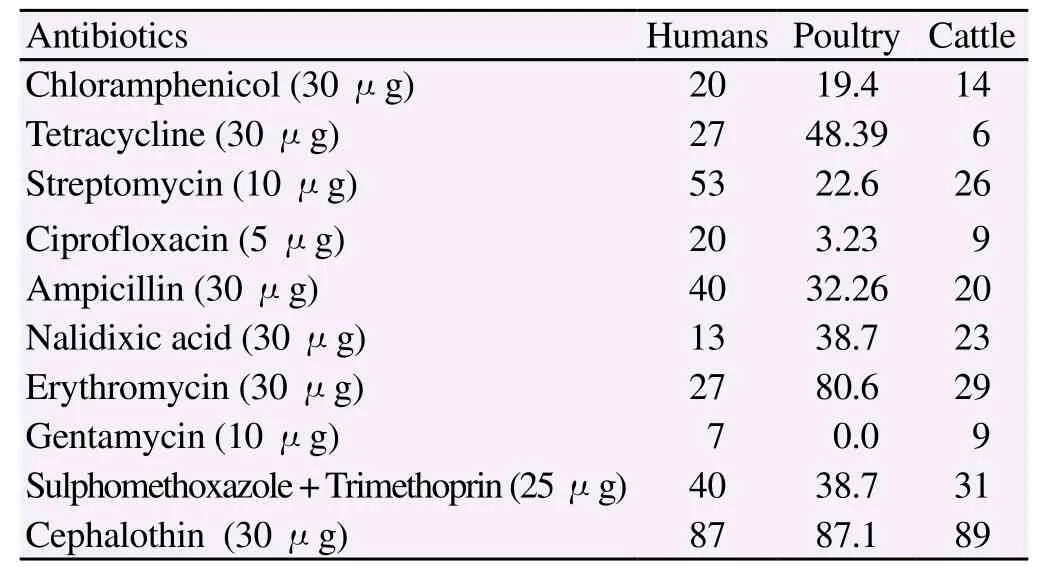
Table 2 Percentages of antibiotic resistances of Campylobacter jejuni isolated from humans, poultry and cattle sources in Pakistan.
Beta-lactamase production was detected in 27 C. jejuni strains including 6 human, 10 poultry and 11 cattle strains. Thus, the overall frequency of beta lactamase producing strains in our study was 27/100 (27%).
3.4. RAPD Profiling
Analysis of C. jejuni isolates by RAPD profiling yielded 22 different banding profiles. Almost all the C. jejuni isolates were well dispersed among all clusters (Figure 2). However, five isolates did not produce any recognizable RAPD banding pattern.
3.5. Virulence typing
Virulence typing was performed using 4 genes as targets and results suggested that cadF and mapA (adherence factors) were present in all isolates studied whereas neuAB (invasive factor) was found in 9 (36%) poultry samples only, whereas wlaN (invasive factor) was not present in any of the isolates (Figure 3).
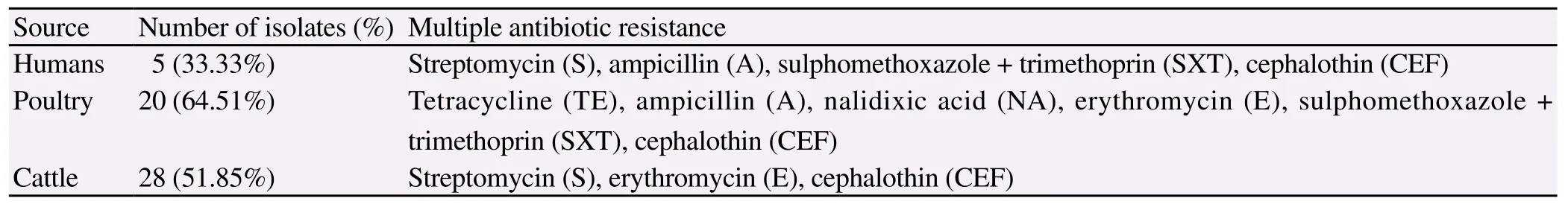
Table 3 Multidrug resistant strains from different sources.
4. Discussion
The aim of this study was to assess the C. jejuni isolates obtained from different sources on the basis of antimicrobial resistance and thereafter screening them for virulence factors. The isolates were further characterized using RAPD analysis for possible relatedness. Little data is available from Pakistan to compare our data, previously isolation rates of C. jejuni from humans have been reported to be 29.5%[21], 12%[22] and 18%[23] and 21.5% Campylobacter spp. prevalence in food commodities[4]. While to our knowledge no reports are available for C. jejuni prevalence in poultry and cattle from Pakistan. The higher prevalence rates 100/436 (22.93%) of
C. jejuni in this study are in agreement with reports from other countries[24-30]. Antibiotic susceptibility profile of C. jejuni isolates was determined using 10 antibiotics and compared among poultry, cattle and humans isolates. The results of antimicrobial susceptibility testing in this study indicate that the isolates were in general resistant to the tested antibiotics at rates ranging from 7% to 87% in clinical cases, up to 87.1% in poultry and 6% to 89% in cattle. Higher rate of resistance (80.6%) to erythromycin was seen among C. jejuni isolates from poultry. Since the ingestion of the infected poultry meat may account for most of human campylobacteriosis cases, this fact becomes more relevant to public health when seen in the context that Erythromycin is one of the commonly used drug for treatment of the patients. However, the frequency of resistance to ampicillin (40%), Tetracycline (27%) and gentamycin (7%) was comparable or lower than in the reports from most of the European countries[31,32]. Mostly tested isolates were susceptible to chloramphenicol and gentamycin. Among the isolates from different sources overall resistance rates were different. Tetracycline was listed as an alternative treatment for Campylobacter gastroenteritis in the past and they are widely used therapeutically and sub therapeutically as feed additives for livestock and poultry[33]. In our study, resistance to tetracycline (7%) was lower than previous reports[34-36]. The identification of multiple antibiotic resistant C. jejuni isolates from poultry, cattle and humans is alarming as such resistance strains may cause more prolonged or severe illness[37]. Further, 27 C. jejuni isolates of during the current study were B-lactamase producers. This is of significance as beta lactams are generally the first line of drugs for treating hospitalized cases. Campylobacter spp. are generally inherently resistant to many beta-lactams, however, there are variable reports of resistance to beta-lactams and some of the expanded spectrum cephalosporins but it is not clearly defined[14]. In our study 6 human diarrheal, 10 poultry and 11 cattle isolates were positive for resistance to B-lactams.
Virulence typing suggested that all isolates possess adherence property owing to the presence of cadF and mapA genes, while 36% of only poultry C. jejuni isolates possess in addition invasive property attributable to the presence of neuAB implying their possibility of association with more severe disease. RAPD typing[38] results have shown the presence of 8 distinct types of C. jejuni. Despite some limitations, analysis of Campylobacter spp. isolates using RAPD has proved to be useful for preliminary characterization of strains[39] and the dendrogram constructed showed genetic diversity of isolatesfrom different sources. Three main clusters were clearly defined ie. clusters Ⅰ, Ⅱ and Ⅲ based on RAPD profiling. All invasive strains (strains positive for neuAB) were present in cluster I whereas all multidrug resistant and beta lactamase producing strains were randomly distributed in all clusters. Our study have shown that RAPD PCR assay can act as rapid and effective molecular tool, which can be used in any basic microbiology laboratory, for studying C. jejuni isolates from different sources and discriminating virulent strains.
This study analyses C. jejuni strains in Pakistani poultry, cattle and human diarrheal samples particularly with regard to their antibiotic resistance and virulence profiling. As compared with European surveillance programmes, the prevalence and antibiotic resistance of C. jejuni in Pakistan are not monitored and isolation of multiple antibiotic resistance C. jejuni from poultry and cattle during the current study serves as impetus for more elaborate studies regarding the prevalence and transmission patterns of C. jejuni.
Acknowledgements
The authors are thankful to British Council for providing funds for this project (Grant SP019) through their strategic partnership awards (INSPIRE Program).
Conflict of interest statement
The authors declare no conflict of interest.
[1] Moore JE, Corcoran D, Dooley JS, Fanning S, Lucey B, Matsuda M, et al. Campylobacter. Vet Res 2005; 36: 351-382.
[2] Silva J, Leite D, Fernandes M, Mena C, Gibbs P, Teixeira P. Campylobacter spp. as a foodborne pathogen: A review. Front Microbiol 2011; 2: 200.
[3] Butzler PJ. Campylobacter, from obscurity to celebrity. Clin Microbiol Infect 2004; 10: 868-876.
[4] Hussain I, Mahmood MS, Akhtar M, Khan A. Prevalence of Campylobacter species in meat, milk and other food commodities in Pakistan. Food Microbiol 2007; 24: 219-222.
[5] Lawes J, Vidal A, Clifton-Hadley F, Sayers R, Rodgers J, Snow L, et al. Investigation of prevalence and risk factors for Campylobacter in broiler flocks at slaughter: results from a UK survey. Epidemiol Infect 2012; 140:1725-37.
[6] Torralbo A, Borge C, Allepuz A, García-Bocanegra I, Sheppard S, Perea A, et al. Prevalence and risk factors of Campylobacter infection in broiler flocks from southern Spain. Prevent Veter Med 2014; 114: 106-113.
[7] Maron D, Smith T, Nachman K. Restrictions on antimicrobial use in food animal production: an international regulatory and economic survey. Globaliz Health 2013, 9:48.
[8] Houndt T, Ochman H. Long-term shifts in patterns of antibiotic resistance in enteric bacteria. Appl Environ Microbiol 2011; 66: 5406-5409.
[9] Taylor DE, Tracz DM. Mechanisms of antimicrobial resistance in Campylobacter. In: Ketley JM, Konkel ME (Eds.). Campylobacter: molecular and cellular biology. Norfolk: Horizon Bioscience. 2005, p. 193-204.
[10] Payot S, Bolla JM, Corcoran D, Fanning S, Megraud F, Zhang Q. Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect 2006; 8: 1967-1971.
[11] Smith J, Fratamico P. Fluoroquinolone resistance in campylobacter. J Food Prot 2010; 73: 1141-1152.
[12] Pollett S, Rocha C, Zerpa R, Patiño L, Valencia A, Camiña M, et al. Campylobacter antimicrobial resistance in Peru: a ten-year observational study. BMC Infect Dis 2012; 12:193.
[13] Luo N, Pereira S, Sahin O, Lin J, Huang S, Michel L, et al. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc Natl Acad Sci USA 2005; 102: 541-546.
[14] Wieczorek K, Osek J. Antimicrobial resistance mechanisms among Campylobacter. Biomed Res Int 2013; 340605.
[15] Aydon F, Atabay HI, Akan M. The isolation and characterization of Campylobacter jejuni subsp. jejuni from domestic geese. J Appl Microbiol 2000; 90: 637-642.
[16] Chaban B, Ngeleka M, Hill JE. Detection and quantification of 14 Campylobacter species in pet dogs reveals an increase in species richness in faeces of diarrheic animals. BMC Microbiol 2010; 10: 1-7.
[17] Cardarelli-Leite P, Blom K, Patton C, Nicholson MA, Steingerwalt AG, Hunter SB, et al. Rapid identification of Campylobacter species strains by Restriction Fragment Length Polymorphism analysis of a PCR-amplified fragment of the gene coding for 16S rRNA. J Clin Microbiol 1996; 34: 62-67.
[18] Atanassova V, und Ring Ch, Nachweis von. Campylobacter spp. mittels RFLP Analyse und PCR Amplifikat Fragment fuer 16S rRNA. In: Deutschland S, editor. 41 Arbeitstagung der Arbeitsgruppe Lebensmittelhygiene der DVG, Garmisch-Partenkirchen. 2000, p. 383-388.
[19] Gaudreau C, Gilbert H. Comparison of disc diffusion and agar dilution methods for antibiotic susceptibility testing of Campylobacter jejuni subsp. jejuni and Campylobacter coli. J Antimicrob Chemother 1997; 39: 707-712.
[20] Lachance N, Gaudreau C, Lamothe F, Turgelon F. Susceptibilities of beta-lactamase positive and -negative strains of Campylobacter coli to beta-lactam agents. Antimicrob Agents Chemother 1993; 37: 1174-1176.
[21] Kazmi RR, Hafeez A, Kazmi SU. Polymicrobial infection in Campylobacter jejuni enteritis in Karachi. FEMS Microb Lett 1987; 41:153-156.
[22] Khalil K, Lindblom GB, Mazhar K. Early child health in Lahore, Pakistan: VII. microbiology. Acta Paediatr 1993; 390: 87-94.
[23] Ali AM, Qureshi AH, Rafi S, Roshan E, Khan I, Malik AM, et al. Frequency of Campylobacter jejuni in diarrhoea/dysentery in children in Rawalpindi and Islamabad. J Pak Med Assoc 2003; 53: 517-520.
[24] Whyte P, McGill K, Cowley D, Madden RH, Moran L, Scates P, et al. Occurrence of Campylobacter in retail foods in Ireland. Int J Food Microb 2004; 95: 111-118.
[25] Ghafir Y, China B, Dierick K, De Zutter L, Daube G. A seven survey of Campylobacter contamination in meat at different production stages in Belgium. Int J Food Microb 2007; 116: 111-120.
[26] Yildirim M, Istanbulluoglu E, Ayvali B. Prevalence and antibiotic susceptibility of thermophilic Campylobacter species in broiler chickens. Turk J Vet Animal Sci 2005; 29: 655-660.
[27] Taremi M, Dallal SMM, Gachkar L, Ardalan MS, Zolfagharian K, Zali, MR. Prevalence and antimicrobial resistance of Campylobacter isolated from retail raw chicken and beef meat, Tehran, Iran. Int J Food Microb 2006; 108: 401-403.
[28] Bostan K, Aydin A, Ang MK. Prevalence and Antibiotic susceptibility of thermophilic Campylobacter species on beef, mutton and chicken carcasses in Istanbul, Turkey. Microb Drug Res 2009; 15: 143-149.
[29] Rahimi E, Ameri M, Kazemeini HR. Prevalence and antimicrobial resistance of Campylobacter species isolated from raw camel, beef, lamb and goat meat in Iran. Foodborne Path Dis 2010; 7: 443-447.
[30] Hassanain N. Antimicrobial resistant Campylobacter jejuni isolated from humans and animals in Egypt. Glob Veterinar 2011; 6: 195-200.
[31] Oporto B, Juste R, Hurtado A. Phenotypic and genotypic antimicrobial resistance profiles of Campylobacter jejuni isolated from cattle, sheep, and free-range poultry faeces. Int J Microbiol 2009. [Online]Available from: http://dx.doi.org/10.1155/2009/456573.
[32] Mattheus W, Botteldoorn N, Heylen K, Pochet B, Dierick K. Trend analysis of antimicrobial resistance in Campylobacter jejuni and Campylobacter coli isolated from belgian pork and poultry meat products using surveillance data of 2004-2009. Foodborn Pathog Dis 2012; 9: 465-472.
[33] Trieber CA, Taylor DE. Mechanisms of antibiotic resistance in Campylobacter. In: Campylobacter. 2nd ed. Washington: ASM Press; 2000, p. 441-454.
[34] Albert M, Udo E, Jose B, Haridas S, Rotimi V. Tetracycline resistance is frequent among Campylobacter jejuni isolates from Kuwait. Microb Drug Resis 2009; 15: 115-120.
[35] Rohini R, Diana S, Harry H, Claude D, Cherie H, Cecelia Y, et al. Fluoroquinolone and metronidazole resistance of Campylobacter spp from broiler chickens and antimicrobial use on farms in Grenada, West Indies. J Anim Res 2012; 2: 219-227.
[36] Wimalarathna H, Richardson J, Lawson A, Elson R, Meldrum R, Little C, et al. Widespread acquisition of antimicrobial resistance among Campylobacter isolates from UK retail poultry and evidence for clonal expansion of resistant lineages. BMC Microbiol 2013; 13: 160.
[37] Traver K, Barza M. Morbidity of infections caused by antimicrobial resistant bacteria. Clin Infect Dis 2002; 34: S131-S134.
[38] Hernandez J, Fayos A, Ferrus MA, Owen RJ. Random amplified polymorphic DNA fingerprinting of Campylobacter jejuni and Campylobacter coli isolated from human faeces, seawater and poultry products. Res Microbiol 1995; 146: 685-696.
[39] Elango A, Dhanalakshmi B, Pugazhenthi T, Jayalalitha V, Kumar C, Doraisamy K. RAPD-PCR characterization of Campylobacter jejuni isolates obtained from raw milk samples in Chennai. Egyp J Dairy Sci 2009; 37: 175-181.
[40] Persson S, Olsen KE. Multiplex PCR for identification of Campylobacter coli and Campylobacter jejuni from pure cultures and directly on stool samples. J Med Microbiol 2005; 54: 1043-1047.
[41] Stucki U, Frey J, Nicolet J, Burnens AP. Identification of Campylobacter jejuni on the basis of a species-specific gene that encodes a membrane protein. J Clin Microbiol 2005; 33: 855-859.
[42] Konkel ME, Gray SA, Kim BJ, Garvis SG, Yoon J. Identification of the enteropathogens Campylobacter jejuni and Campylobacter coli based on the cadF virulence gene and its product. J Clin Microbiol 1999; 37: 510-517.
[43] Parker CT. Diversity in the lipooligosaccharide biosynthesis locus of Campylobacter jejuni. NCBI accession number AY434498. Produce Safety and Microbiology Unit, United States Department of Agriculture, Agriculture Research Service, Albany, Calif. 2004.
[44] Wassenaar TM, Wagenaar JA, Rigter A, Fearnley C, Newell DG, Duim B. Homonucleotide stretches in chromosomal DNA of Campylobacter jejuni display high frequency polymorphism as detected by direct PCR analysis. FEMS Microbiol Lett 2002; 212: 77-85.
ent heading
10.1016/S1995-7645(14)60314-X
*Corresponding author: Prof. Dr. Habib Bokhari, Chairman, Department of Biosciences, COMSATS Institute of Information Technology, Park Road, 44000, Islamabad, Pakistan.
Tel: 00923005127684
Fax: 0092214442805
E-mail: habib@comsats.edu.pk
 Asian Pacific Journal of Tropical Medicine2015年3期
Asian Pacific Journal of Tropical Medicine2015年3期
- Asian Pacific Journal of Tropical Medicine的其它文章
- Statistical estimations for Plasmodium vivax malaria in South Korea
- Evolutionary relationship of 5’-untranslated regions among Thai dengue-3 viruses, Bangkok isolates, during 24 year-evolution
- Depressant effects of Agastache mexicana methanol extract and one of major metabolites tilianin
- Fatty acid methyl ester profiles and nutritive values of 20 marine microalgae in Korea
- Prevalence of West Nile virus in Mashhad, Iran: A population-based study
- Effects of high glucose on expression of OPG and RANKL in rat aortic vascular smooth muscle cells
