Characterization of the Genetic Diversity of Trachemys dorbigni and Phrynops hilarii
GUIDETTI Brenda Yamile, SIROSKI Pablo Ariel and AMAVET Patricia Susana
Facultad de Humanidades y Ciencias, Universidad Nacional del Litoral, 3000 Santa Fe, Argentina
Characterization of the Genetic Diversity of Trachemys dorbigni and Phrynops hilarii
GUIDETTI Brenda Yamile*, SIROSKI Pablo Ariel and AMAVET Patricia Susana
Facultad de Humanidades y Ciencias, Universidad Nacional del Litoral, 3000 Santa Fe, Argentina
The utilization of RAPD and ISSR molecular markers is proposed to initiate studies of genetic variability in Phrynops hilarii (Chelidae) and Trachemys dorbigni (Emydidae), two species of fresh water turtles distributed in South America. Three primers of RAPD and four of ISSR were selected and the amplified products of these markers were evaluated by electrophoretic runs in agarose and polyacrylamide gels. The levels of heterozygosity, Shannon index and different allele numbers were slightly higher in P. hilarii for both types of markers. Levels of polymorphism were also higher in P. hilarii than T. dorbigni and both were elevated compared to those recorded for other species. The fact that similar results were obtained with both types of markers for all estimates of diversity highlights the usefulness and validity of the RAPD technique. The molecular markers used were found potentially useful for analysing future temporal and spatial distribution of genetic diversity in both species, expanding scales work.
fresh water turtles, variability, molecular markers, RAPD, ISSR
1. Introduction
According to the method of Hoffmann et al. (2010), between 48% and 54% of turtle species in the world are threatened or endangered. Longevity and overlapping generations that characterize the group mask the problem, with severe demographic effects which are not yet observable likely to take years or decades to manifest (Fagundes et al., 2010). Some turtles may exhibit little plasticity in habitat use, in freshwater turtles this can be particularly serious because they generally require more than one environment throughout the life cycle. Despite this, at least in Argentina, those species receive the least protection within the group (Ubeda and Grigera, 2003). Trachemys dorbigni (Emydidae) and Phrynops hilarii (Chelidae) are freshwater turtles widely distributed in South America. Neither of the two species are considered endangered (CITES, 2011; IUCN, 2011). Phrynops hilarii is well represented across its vast distribution, even more, the area of the species has expanded inrecent times favoured by anthropochory, showing the ability to adapt to highly modified habitats (Richard, 1999). On the other hand, according to the evaluation index for tetrapod fauna proposed by Reca et al. (1994), T. dorbigni can be considered a vulnerable species and there are many reasons for the current situation to be aggravated: the species is being affected by the advance of the agricultural frontier, is capable of hybridizing with native species (Lavilla et al., 2000; Richard, 1999), and also suffers a severe pressure through egg and juvenile capture for illegal trafficing that supplies pet markets and has high levels of exploitation for consumption of meat and meat products (Bujes, 2010; Carreira et al., 2007; Fagundes et al., 2010). What is more, both species live in environments with polluted waterways, where there also have been established hydroelectric stems (Bujes, 2010). Ecologists have also started to become aware of the many deaths resulting from accidents on roads and paths (Bager et al., 2007; Bujes, 2010).
The necessity to update the information referring to population parameters, current distribution and conservation status of these species motivates the realization of this study. Mostly, T. dorbigni genetic studies are related to karyotypic analysis and cytogenetictechniques (Martinez et al., 2009; Salas, 2011) and as far as is known, there are no genetic studies for P. hilarii. This lack of antecedents was considered sufficient to initiate studies of the genetic variability by comparing the inter and intra specific diversity. It was proposed to employ the methodology of two molecular DNA markers: RAPD (random amplified polymorphic DNA) and ISSR (intersimple sequence repeats ), that have been used successfully in other species of aquatic turtles (Duan et al., 2011; Zheng et al., 2008; Zhu et al., 2008; Zhu, 2011). Both techniques are practical, simple, easily reproducible and cheap, which allows researchers to obtain results quickly and without destructive or highly invasive sampling for a large number of individuals (Rentaria Alcántara, 2007; Rocha and Gasca, 2007; Zietkiewicz et al., 1994). Considering broader scales of work, such methodologies allowed one to obtain a large amount of genetic information for analysing the temporal and spatial distribution of genetic diversity of species, which is the foundation for planning accurate actions for species conservation (Alacs et al., 2007; Ma et al., 2007b; Souza et al., 2002; Zhu et al., 2005). These molecular markers can also be useful to determine the origin of organisms extracted from their habitat, to reintroduce them at appropriate locations (Rocha and Gasca, 2007) or guide the management of captive populations in reserves, zoos and urban areas (Amavet et al., 2009).
2. Methods
2.1 Sample collection and DNA extraction Twenty six (26) blood samples were obtained from adult specimens (Phrynops hilarii, N = 13 and Trachemys dorbigni, N = 13) from the Applied Zoology Laboratory: Vertebrates (MASPyMA / FHUC UNL) according to Olson’s technique (Olson et al., 1975). All the individuals used in this experiment were rescued during seizure operations and the original geographical location of each specimen is unknown. DNA extraction was performed using the technique of extraction of Murray and Thompson (1980) from blood samples diluted (1:10) in a lysis buffer (Longmire et al., 1988) for long-term blood storage at room temperature according to White and Densmore (1992). The extractions for each individual were stored at 4 ± 2°C until to test the quality and quantity of extracted DNA by electrophoresis using 0.8% agarose gels, runs at 120 V in 0.5× TBE (Tris/Borate/EDTA) buffer, stained with Gel Green (Biotium) and analysed in dark light transilluminator (Dark Reader). DNA samples were diluted with H2O 1:4, 1:3 and mostly 1:2.
2.2 Amplifcation of polymorphic regions with primers RAPD and ISSR Three samples for each species were used to screen a set of 10 primers from Promega® (B050-10 and B051-10) for RAPD and 13 primers from Operon® series for ISSR, to test amplification profiles for readability and reproducibility. Seven primers that showed the best resolution and reproducible bands were selected to obtain RAPD and ISSR profiles for all individuals, and three repeatability test samples were included in each amplification reaction.
The RAPD amplification reactions were carried out at first in accordance to the methodology of Bardakci and Skibinski (1994), in a final volume of 15 μl, containing 1.5 μl of buffer, 1.5 μl of dATP, dTTP, dGTP and dCTP solution (200 mM), 0.75 μl of the selected primer, 1.5 μl of MgCl2, 0.15 μl of Taq DNA polymerase (PB-L®)and 50 ng of genomic DNA. Occasionally, the amounts of reactants were adjusted, such as varying the amount of primer from 0.75-1 μl and then from 1-1.2 μl. The amount of Taq DNA polymerase (PB-L®) must also be increased from 0.15-0.2 μl, but returned to settle in 0.15 μl when it was decided to use the brand Invitrogen®. Amplifications were performed in a thermocycler (MPI®)with a program of 40 cycles of 1 min at 94°C, 1 min at 40°C and 1 min at 72°C, with an initial denaturation of 94°C for 4 minutes and a final extension at 72°C for 10 minutes.
To amplify ISSR regions, reactions were performed to a final volume of 15 μl containing 1.5 μl buffer, 1 μl dATP , dTTP , dGTP and dCTP solution (200 mM),1 μl of the selected primer, 1 μl of MgCl2, 0.15 μl of Taq DNA polymerase (PB-L®) and 50 ng of genomic DNA. Amplifications were performed in a thermocycler (MPI®) with a program of 40 cycles of 1 min at 94°C, 1 min at annealing temperature according to the selected primer and 1 min at 72°C , with an initial denaturation of 94°C for 4 minutes and a final extension at 72°C for 10 minutes.
2.3 Analysis of markers For the selection of primers, the PCR products were visualized and analyzed by electrophoretic runs in 2% agarose gels, at 120–130 V in 0.5× TBE buffer. For RAPD: 3 primers Series A were selected (Table 1) and the PCR products corresponding to these primers were analyzed by vertical electrophoresis runs performed in 4% polyacrylamide gels, of 33 cm × 39 cm, run at 220 V and 75 W in 0.5× TBE buffer for 2:30 to 3 hours, with a 30 minutes pre-electrophoretic run. Gel staining was performed with silver nitrate, using the methodology of Bassam et al. (1991) described by Promega®. In all electrophoresis runs, DNA ladders (10bp from Invitrogen® and 100 bp from PB-L®) were used to estimate the size of the amplified fragments. For ISSR:4 primers were selected (Table 1) and the PCR products were analyzed by electrophoresis runs performed in 6% non-denaturing polyacrylamide gels, of 10 cm × 10 cm, at 120V–130V in 0.1× TBE buffer. Pre-electrophoretic run was performed during 20 minutes, and finally the gel was electrophoresed for period between 2 and 2:40 hours. The staining was performed with silver nitrate, using the methodology of Herring et al. (1982). A molecular weight marker (O’RangeRuler®20 bp DNA Ladder) was used in all electrophoresis runs to estimate the size of the amplified fragments. All gels were observed with background light and photographed with an Olympus® C- 5000 Zoom 5.0 Megapixel digital camera.
2.4 Data Analysis The bands are interpreted as present if they can be clearly detected, whether they had more or less intensity. Through the observation of all obtained bands in the gels, we built binary matrices that were analyzed using the program GenAlEx (version 6.41) (Peakall and Smouse, 2006).
Measures of genetic variability thrown by these program for each species were: original (1972) and unbiased Nei genetic distance (1978), percentage of polymorphic loci = number of polymorphic loci/total number of loci analyzed, He = heterozygosity expected (on HW equilibrium) = 2 * p * q; UHE = unbiased heterozygosity (Nei) = (2N / (2N-1)) * I ; I = Shannon’s information index = –1 * (p * Ln (p) + q * Ln (q)) (used in ecology to measure the specific biodiversity and considered robust for dominant markers when heterozygous loci cannot be detect); Na = Number of alleles and Ne = number of effective alleles = 1 / (p2+ q2). Keep in mind that for diploid binary data, Hardy-Weinberg equilibrium is assumed: q = (1 – frequency band) 0.5 and p = 1 – q.
3. Results
The total number of loci (bands) analyzed with the 3 RAPD primers was 122, with an average of 40.66 per primer. Of the 122 bands, 104 were amplified in individuals of P. hilarii and 102 in individuals of T. dorbigni, all at a higher frequency than 5%. Numerous bands that appear only in one species were observed (20 exclusive bands of P. hilarii and 18 bands in T. dorbigni), but these bands were not recorded in all individuals of the species. The total number of amplified fragments was 866, considering 439 fragments in P. hilarii and 427 for T. dorbigni, with an average number of fragments per individual of 33.77 for P. hilarii and 32.85 for T. dorbigni. The size of the PCR products ranged from 352 bp to about 2419 bp.
The total number of loci (bands) analyzed with the 4 ISSR primers was 117, with an average of 29.25 per primer. Of the 117 bands, 106 amplified in individuals of P. hilarii and 86 in individuals of T. dorbigni, all at a higher frequency than 5%. None of the 31 exclusive bands of P. hilarii found occurred in all individuals, on average these exclusive bands appeared only in 3.48 individuals. In T. dorbigni, only one of 11 unique bands observed is presented in all sampled individuals (average exclusive bands appear in 5.09 individuals). The total number of fragments amplified with 4 primers of ISSR was 1040, considering 470 fragments for P. hilarii and 570 for T. dorbigni. The average number of fragments per individual was 36.15 for P. hilarii and 43.85 for T. dorbigni. The descriptive statistics values (percentage of polymorphic loci, genetic distance, expected heterozygosity, unbiased heterozygosity, Shannon index, different alleles and effective alleles) obtained in both species by analysis of RAPD and ISSR markers employing software GenAlEx are summarized in Tables 2 and 3.
The percentage of polymorphic loci obtained in this study with RAPD were similar for both species (84.43% for Phrynops hilarii and 82.79% for Trachemys dorbigni) and could be considered high when compared to those found for other species studied with these types of molecular markers (Duan et al., 2011; Ma et al., 2007a; Zheng et al., 2008; Zhu, 2011). The estimated percentage of polymorphic loci based on ISSR was higher in P. hilarii (89.74%) than in T. dorbigni (69.23%). These percentages reaffirm the idea that levels of polymorphism within Chelonia are not as low as previously thought (Souza et al., 2002). It is also true that the estimates of polymorphism could be sensitive to a certain level of subjectivity during the counting of the bands, and somehow this would limit the possibility of making comparisons between different studies.
Levels of expected heterozygosity (He) and unbiased heterozygosity (UHE) were similar in both species, slightly higher in P. hilarii when compared to T. dorbigni for RAPD and ISSR markers. The values from the Shannon information index (I) (0.348 ± 0.021 in P. hilarii and 0.327 ± 0.020 in T. dorbigni with RAPD, and 0.379 ± 0.019 in P. hilarii and 0.345 ± 0.026 in T. dorbigni with ISSR) and the number of different alleles (1.697 ± 0.065 and 1.664 ± 0.067 with RAPD and 1.803 ± 0.055 and 1.427 ± 0.082 with ISSR for P. hilarii and T. dorbigni, respectively) are also similar in both species, barelyhigher in P. hilarii than T. dorbigni for both marker types. Only with ISSR the number of effective alleles per locus is somewhat higher in T. dorbigni (1.407 ± 0.037, in contrast with 1.370 ± 0.027 for P. hilarii). The values of the descriptive statistics obtained with RAPD and ISSR molecular markers for P. hilarii are higher in all cases comparing to those obtained for T. dorbigni, suggesting more genetic variability in this species.
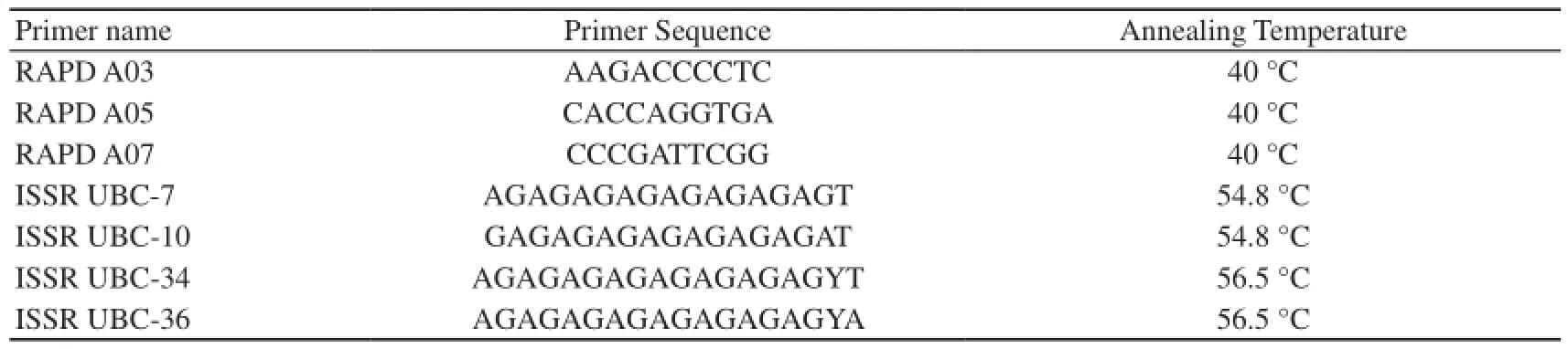
Table 1 RAPD (Promega®) and ISSR (Operon®) primers selected to perform PCR amplifications.
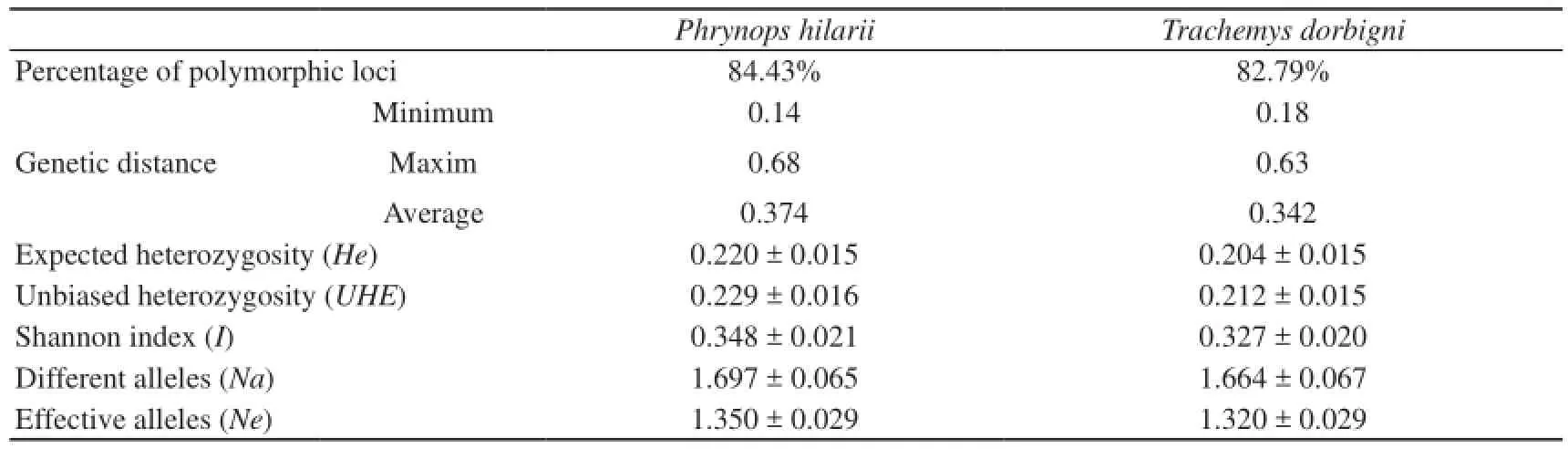
Table 2 Descriptive statistics obtained with RAPD primers according to GenAlEx software.
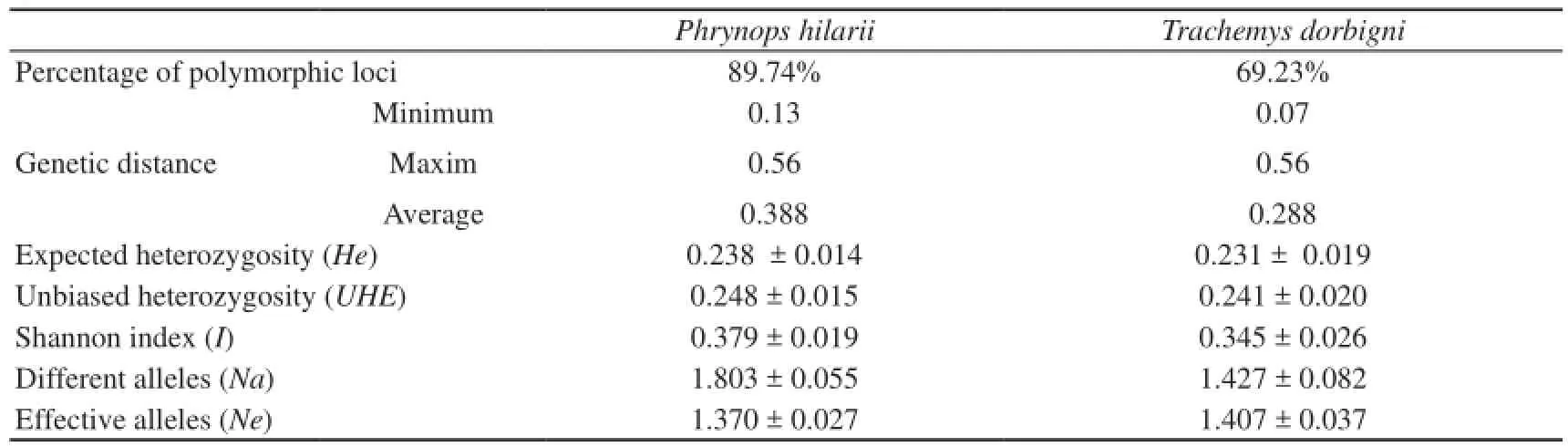
Table 3 Descriptive statistics obtained with ISSR primers according to GenAlEx software.
4. Discussion
Molecular methodologies used in this study proved to be effective for an initial screening of the genetic variability of these species. We were able to observe a higher number of variable markers than other similar studies in turtles. The total number of loci (bands) analyzed was 117 with the four ISSR primers and 122 with the 3 RAPD primers. In Semyenova et al. (2004) the five RAPD primers allowed amplification of a total pool containing 180 fragments, while Zhu et al. (2008) scored a total of 20 population-specific RAPD fragments from 16 primers. In Zheng et al. (2008) 8 fragments were obtained with each of the 12 RAPD primers. This would be due in part to the use of polyacrylamide gels, which provide greater resolution and depth analysis. These methods also allow working with small amounts of DNA and it is not necessary to have prior knowledge of their sequence, furthermore, nonradioactive probes are required in the process (Grosberg et al., 1996; Lynch and Milligan, 1994; Rocha and Gasca, 2007).
In both species for RAPD and ISSR, the numbers of different alleles were higher than the effective numbers of alleles, which might suppose that the presence of low frequency alleles may influence the presenceof heterozygous, causing a decrease in the genetic variability.
The RAPD genetic distance calculated between individuals of the same species showed peak (0.140–0.680 for P. hilarii; 0.180–0.630 for T. dorbigni) and mean values (0.374 for P. hilarii; 0.342 for T. dorbigni) that may be somewhat high if one takes into account those recorded for other species studied with RAPD. Duan et al. (2011) calculate genetic distances from 0.0829 to 0.1813 and an average of 0.1327 ± 0.0299 in Eretmochelys imbricate. For Chinemys reevesii the genetic distance calculated ranged from 0.168 to 0.467, and the average was 0.324 ± 0.0631 according to Zhu et al. (2005) but ranged from 0.1360 to 0.3609, with an average of 0.2092 ± 0.0623 according to Zhu (2011). In Mauremys mutica the average genetic distance among two populations was 0.299 ± 0.108 (Zhu et al., 2008). The genetic distance between individuals of the two species determined by ISSR markers in this study also showed a wide range with high maximum values (0.130–0.560 for P. hilarii; 0.560–0.070 for T. dorbigni). These high values suggest that individuals sampled may belong to different populations, situated in distant geographical locations (the exact origin of the rescued individuals is unknown, reason for which this statement cannot be tested).
The RAPD and ISSR markers may be useful to analyse the temporal and spatial distribution of genetic diversity in these species. Most amphibians and reptiles exhibit deep phylogeographic differentiation, basically due to their low vagility, understood as the distance between the point of birth of an individual and the point of death (constituting a parameter that define mobility or dispersion, referring to the ability or tendency of individuals or populations to spread, changing its distribution over time) (Fagundes et al., 2010; Souza et al., 2002). Based on site fidelity, reduced dispersal and longevity, turtles seem to be an interesting group to include in phylogenetic studies. Moreover, high levels of population structure were recognized, it would be possible and necessary to begin delineating ESUs (Evolutionarily Significant Units) that consider and protect long-term evolutionary potentials (Vázquez Domínguez, 2007).
The ISSR technique is very useful to evaluate diversity in species. Given the high genetic variation between individuals within a population, it is possible to use these markers in paternity analysis and identification of individuals (Rentaría Alcantara, 2007; Rocha and Gasca, 2007). Their high polymorphism also allows to apply them in distinction of intraspecific varieties and population genetic subdivision, including complex cases where gene flow, introgression and hybridization is evidenced (Ma et al., 2007b, Schilde et al., 2004). They have also been used to perform genetic mapping and phylogenetic reconstruction including sometimes cryptic species distinction (Fritz et al., 2005, 2007).
Given the high polymorphism detect by RAPDs, they have proven to be useful in the genetic identification (including clones, hybrid or mutant) and the study of relationship. The technique is also applied in genetic mapping, detection of genetic uniformity and analysis of intraspecific population structure at different spatial scales, allowing estimating effective size, reproductive isolation and levels of crossing fecundation (Alacs et al., 2007; Rocha and Gasca, 2007; Rubin et al., 2001). The RAPD markers facilitate the realization of fast and efficient analysis of genetic variability in not well known,vulnerable or endangered species, also in those that are of economic interest to subsistence of certain societies (Duan et al., 2011; Ma et al., 2007a; Mockford et al., 1999; Tan et al., 2000; Zheng et al., 2008; Zhu et al., 2008). The genetic information collected usually can be integrated with ecological data in advance, allowing the development of more effective conservation strategies (Souza et al., 2002).
5. Conclusions
Estimators of genetic variability for Phrynops hilarii and Trachemys dorbigni were obtained from the use of molecular markers RAPD and ISSR, two relatively new, simple, fast and economical techniques, so far had not been used in any of the two species under study. It is also considered that the use of polyacrylamide gels increases the resolution of the analysis bands.
The usefulness and validity of RAPD is reinforced in this study by the fact that very similar results were obtained with both types of markers for all diversity estimators. Taking the necessary precautions (appropriate laboratory conditions, negative controls and repetitions, etc.) the lack of reproducibility that sometimes has been criticized for this technique can be avoided.
The results achieved encourage research in both species according to all the possibilities offered by these markers in relation to the lack of studies on them. The information obtained in this work can be useful as a starting point for phylogeographic studies at the population and/or specific level in both P. hilarii and T. dorbigni, suitable for the development of appropriate management strategies to protect and conserve these species in the region.
Acknowledgements We thank the staff of Laboratory of Applied Zoology: Vertebrates, MASPyMA / FHUC-UNL that took blood samples and made available for this work.
Alacs A., Janzen F., Scribner K. 2007. Genetic issues in freshwater turtle and tortoise conservation. In Shaffer H., FitzSimmons N., Georges A., Rhodin A. (Eds.), Defining Turtle Diversity: Proceedings of a Workshop on Genetics, Ethics, and Taxonomy of Freshwater Turtles and Tortoises. Chelonian Research Foundation, Chelon Res Monogr, 4: 134–152
Amavet P., Vilardi J., Rosso E., Saidman B. 2009. Genetic and morphometric variability in Caiman latirostris (broad-snouted caiman), reptilia, alligatoridae. J Exp Zool, 309A: 1–12
Bager A., de Freitas T., Krause L. 2007. Nesting ecology of population of Trachemys dorbignyi (Emydidae) in Southern Brazil. Herpetologica, 63(1): 56–65
Bardakci F., Skibinsi O. 1994. Application of the RAPD technique in tilapia fish: Species and subspecies identification. Heredity, 73: 117–123
Bassam B., Caetano-Anollés G., Gresshoff P. 1991. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem, 196: 80–83
Bujes C. 2010. Os Testudines continentais do Rio Grande do Sul, Brasil: Taxonomía, historia natural e conservação. Iheringia, Sér Zool, 4: 413–424
Carreira S., Estrades A., Achaval F. 2007. Estado de conservación de la fauna de tortugas (Reptilia, Testudines) de Uruguay. Boletín Sociedad Zoológica del Uruguay, 16: 20–25
CITES 2011. Convention on International Trade in Endangered Species of Wild Fauna and Flora. Appendix I, II and III (http:// www.cites.org/eng/app/appendices.shtml)
Duan J., Gu H., Xia Z., Ye M., Chen H., Zhang F. 2011. Genetic diversity analysis of Eretmochelys Imbricata by RAPD method. Chin J Wildl, 5: 264–266, 292
Fagundes C., Bager A., Zanini S. 2010. Trachemys dorbigni in an anthropic environment in southern Brazil: Sexual size dimorphism and population estimates. Herpetol J, 20: 185–193
Fritz U., Fattizzo T., Guicking D., Tripepi S., Pennisi., M., Lenk P., Joger U., Wink M. 2005. A new cryptic species of pond turtle from southern Italy, the hottest spot in the range of the genus Emys (Reptilia, Testudines, Emydidae). Zool Scr, 34: 351–371
Fritz U., Hundsdörfer A., Široký P., Auer M., Kami H., Lehmann J., Mazanaeva L., Türkozan O., Wink M. 2007. Phenotypic plasticity leads to incongruence between morphology-based taxonomy and genetic differentiation in western Palaearctic tortoises (Testudo graeca complex; Testudines, Testudinidae). Amphibia-Reptilia, 28: 97–121
Grosberg R., Levitan D., Cameron B. 1996. Characterization of genetic structure and genealogies using RAPD-PCR markers: A random primer for the novice and nervous. In Ferraris J., Palumbi S. (Eds.), Molecular Zoology: Advances, Strategies, and Protocols. New York: Wiley-Liss, 67–100
Herring A., Inglis N., Ojeh C., Snodgrass D., Merizies J. 1982. Rapid diagnosis of rotavirus infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels. J Clin Microbiol, 16: 473–477
Hoffmann M. et al. 2010. The Impact of Conservation on the Status of the World’s Vertebrates. Science, 330: 1503–1509
IUCN. 2011. International Union for Conservation of Nature and Natural Resources. Red List of Threatened Species. Version 2011.1 (http://www.iucnredlist.org)
Lavilla E., Richard E., Scrocchi G. 2000. Categorización de los Anfibios y Reptiles de la República Argentina. Asociación Herpetológica Argentina, San Miguel de Tucumán
Longmire J., Lewis A., Brown N., Buckingham J., Clark L.,Jones M., Meincke L., Meyne J., Ratliff R., Ray F., Wagner R., Moyzis R. 1988. Isolation and molecular characterization of a highly polymorphic centromic tandem repeat in the family Falconidae. Genomics, 2: 14–24
Lynch M., Milligan B. 1994. Analysis of population genetic structure with RAPD markers. Mol Ecol, 3: 91–99
Ma L., Zheng G., Zhu X., Liu Y., Chen Y., Luo J. 2007a. Genetic diversity analysis of Platysternon megacephalum by RAPD method. Freshw Fish, 2: 76–79
Ma L., Zheng G., Zhu X., Liu Y., Chen Y., Luo J. 2007b. Genetic diversity in two natural populations of Platysternon megacephalum as revealed by ISSR technique. Chin J Zool, 49(1): 13–20
Martinez P., Boeris J., Sánchez J., Pastori M., Bolzán A., Ledesma M. 2009. Karyotypic characterization of Trachemys dorbigni (Testudines: Emydidae) and Chelonoidis (Geochelone) donosobarrosi (Testudines: Testudinidae), two species of Cryptodiran turtles from Argentina. Genetica, 3: 277–283
Mockford S, Snyder M., Herman T. 1999. A preliminary examination of genetic variation in a peripheral population of Blanding’s turtle, Emydoidea blandingii. Mol Ecol, 8(2): 323–327
Murray M., Thompson W. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res, 8: 4321–4325
Nei M. 1972. Genetic distance between populations. Amer Nat, 106(949): 283–292
Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89(3): 583–590
Olson G., Hessler J., Faith R. 1975. Technics for blood collection and intravascular infusion of reptiles. Lab Anim Sci, 6: 783–786
Peakall R., Smouse P. 2006. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes, 6: 288–295
Reca A., Úbeda C., Grigera D. 1994. Conservación de la fauna de tetrápodos. Un índice para su evaluación. Mastozool Neotrop, 1(1): 17–28
Rentaría Alcántara M. 2007. Breve revisión de los marcadores moleculares. In Eguiarte L., Souza V., Aguirre X. (Eds.), Ecología molecular. México: Semarnat-UNAM-Conabio, 541–566
Richard E. 1999. Tortugas de las Regiones Áridas de Argentina. Editorial LOLA, Buenos Aires
Rocha M., Gasca J. 2007. Ecología molecular de la conservación. In Eguiarte L., Souza V., Aguirre X. (Eds.), Ecología molecular. México: Semarnat-UNAM-Conabio, 253–278
Rubin C., Warner R., Bouzat J., Paige K. 2001. Populationgenetic structure of Blanding´s turtle (Emydoidea blandingii) in an urban landscape. Biol Cons, 99: 323–330
Salas A. 2011. Estudios citogenéticos en la tortuga pintada (Trachemys dorbigni: Reptilia, Emydidae). Bachelor Thesis UNL-FHUC, Santa Fe, Argentina
Schilde M, Barth D., Fritz U. 2004. An Ocadia sinensis x Cyclemys shanensis hybrid (Testudines:Geoemydidae). Asiat Herpetol Res, 10: 120–125
Semyenova S., Korsunenko A., Vasilyev V., Pereschkolnik S., Mazanaeva L., Bannikova A., Ryskov A. 2004. RAPD variation in Mediterranean turtle Testudo graeca (Testudinidae). Russ J Genet, 12: 1348–1355
Souza F., Cunha A., Oliveira M., Pereira G., Pinheiro H. F. dos Reisa S. 2002. Partitioning of molecular variation at local spatial scales in the vulnerable neotropical freshwater turtle, Hydromedusa maximiliani (Testudines, Chelidae): Implications for the conservation of aquatic organisms in natural hierarchical systems. Biol Cons, 104: 119–126
Tan S., Ng Y., Joseph J., Chan E. 2000. Genetic variation in hawksbill turtle (Eretmochelys imbricata) from Malaysia using RAPD markers. Towards sustainable management of the Straits of Malacca, 261–266
Ubeda C., Grigera D. 2003. Analysis of the last assessment of conservation status of amphibians and reptiles from Argentina. Gayana, 67(1): 97–113
Vázquez Domínguez E. 2007. Filogeografía y vertebrados. In Eguiarte L., Souza V., Aguirre X. (Eds.), Ecología molecular. México: Semarnat-UNAM-Conabio, 441–466
White P., Densmore L. 1992. Mitochondrial DNA isolation. In Hoelzel A. (Eds.), Molecular genetic analysis of populations. A practical approach. The practical approach series. Oxford: Oxford University Press, 29–57
Zheng G., Ma L., Zhu X., Liu Y., Chen Y., Luo J. 2008. Genetic diversity analysis between two populations of Platysternon megacephalum by RAPD. J Huazhong Agric Univ, 27(4): 510–514
Zhu X., Du H., Zhou L., Li M., Gui J. 2005. Genetic diversity analysis of Chinese three-keeled pond turtle (Chinemys reevesii) by RAPD. Acta Hydrobiol Sin, 29: 167–171
Zhu X., Zhou L., Chen Y., Du H., Gui J. 2008. Phenotypic and genetic variation between two populations of the Chinese yellow pond turtle, Mauremys mutica (Cantor, 1842). Chin High Tech Lett, 14: 104–111
Zhu X. 2011. Analysis of genetic diversity amongst Chinemys reevesii in Guangxi using RAPD markers. J South Agric, 42(9): 1148–1150
Zietkiewicz E., Rafalski A., Labuda D. 1994. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics, 20: 176-183
GUIDETTI Brenda Yamile, from Universidad Nacional del Litoral, Santa Fe, Argentina, with his research focusing on conservation ecology.
E-mail: guidettibrenda@gmail.com
7 September 2014 Accepted: 16 March 2015
 Asian Herpetological Research2015年3期
Asian Herpetological Research2015年3期
- Asian Herpetological Research的其它文章
- A New Record of Kaloula (Amphibia: Anura: Microhylidae) in Shanghai, China
- First Record of Male Combat in a Wild Malayan Pit Viper(Calloselasma rhodostoma)
- Reevaluation of the Taxonomic Status of a Poorly Known Gecko, Gekko liboensis (Reptilia: Squamata)
- Acoustic Characteristics of Advertisement Calls in Babina adenopleura
- Potential Distribution Modeling and Morphology of Pelias barani (Böhme and Joger, 1983) in Turkey
- Genetic and Morphological Variations within Laudakia microlepis (Blanford, 1874) (Sauria: Agamidae) Populations in Southeastern Iran with Description of a New Subspecies
