Establishmentand Comparison of Two TaqMan Real-time PCR Methods for PCV2
Zhongtao ZHOU,Xiaomin WANG,Wei WANG,Aihua MAO,Libin WEN,Yanxiu NI,Kongwang HE*
1.Ministry of Agriculture’s Key Laboratory of Veterinary Biologicals Engineering and Technology,National Center for Engineering Research of Veterinary Biological Products,Institute of Veterinary Medicine,Jiangsu Academy of Agricultural Sciences,Nanjing 210014,China;
2.College of Veterinary Medicine,Nanjing Agricultural University,Nanjing 210095,China
Establishmentand Comparison of Two TaqMan Real-time PCR Methods for PCV2
Zhongtao ZHOU1,2,Xiaomin WANG1,Wei WANG1,Aihua MAO1,Libin WEN1,Yanxiu NI1,Kongwang HE1*
1.Ministry of Agriculture’s Key Laboratory of Veterinary Biologicals Engineering and Technology,National Center for Engineering Research of Veterinary Biological Products,Institute of Veterinary Medicine,Jiangsu Academy of Agricultural Sciences,Nanjing 210014,China;
2.College of Veterinary Medicine,Nanjing Agricultural University,Nanjing 210095,China
[Objective]In this study,the quantitive detection of PCV2(porcine circovirus type 2)in vitro was achieved.We aimed to establish two kinds of Taq Man real-time PCR methods based on PCV2 ORF1 and ORF2 respectively and compare them.[Method]According to the relatively conserved sequences of PCV2 ORF1 and ORF2 registered in GenBank,two pairs of specific primers and Taq Man probes were designed and synthesized.Then the recombinant plasmids containing the whole sequences of PCV2 ORF1 and ORF2 were constructed to draw the standard curves through optimizing the reaction system and conditions.And thus two kinds of Taq Man real-time PCR detection methods based on the whole sequences of ORF1 and ORF2 respectively were constructed for PCV2.[Result]For the two established standard curves,the Ct values showed a good linear relationship with the logarithms of copy numbers of templates(R2>0.99).The amplification efficiency ranged from 90%to 110%.The amplifications all had a good repeatability with variation coefficients within groups all less than 5%.Moreover,the amplifications all had a good specificity.When the sequences of porcine parvovirus(PPV),porcine circovirus type 1(PCV1),swine pseudorabies virus(PRV),porcine reproductive and respiratory syndrome virus(PRRSV)were used as templates,the target sequence was not amplified.The amplifications also had a high sensitivity.The ORF1 detection method could reach 1.0×101copies/μl,and the ORF2 detection method could reach 1.0×102copies/μl.The two established real-time PCR detection methods were used to detect the 80 clinical samples respectively.The results showed the magnitudes of 72 amplified samples were basically consistent between the 2 detection methods,while the magnitudes of the other 8 amplified samples were inconsistent.Then the 8 samples were detected with SYBR Green I real-time PCR method established based on the sequence of PCV2-like factor P1 by Wen et al.The PCV2-like factor P1 was amplified in all the 8 samples,indicating the 8 samples were all infected with PCV2-like factor P1.[Conclusion]The ORF1-based detection method has a higher accuracy, and it can be used for the rapid detection of PCV2.
PCV2;TaqMan real-time PCR;ORF1;ORF2;PCV2-like factor P1
2种猪圆环病毒2型TaqMan实时荧光定量PCR检测方法的建立及比较
P orcine circovirus type 2(PCV2) (Circoviridae:Circovirus)was firstly discovered by Tischer et al.[1]in the passaged celllines ofPK-15 in 1974.It is a kind of small singlestranded circular DNA virus without envelope.Allan[2-3]and Brian[4]et al divided the PCV into 2 genotypes, PCV1 and PCV2,according to the pathogenicity,antigenicity and nucleotide sequence.PCV2 can lead to the post-weaning multisystemic wasting syndrome,as wellas porcine dermatitis and nephropathy syndrome, porcine proliferative and necrotizing pneumonia,porcine respiratory syndrome,porcine congenital tremor,reproductive disorders and other diseases,bringing huge economic losses to world pig industry[5-7].
In the detection of PCV2 in samples possibly infected with PMWS by PCR,Wen et al.[8-10]found the factors that are related to the gene sequence of PCV2 ORF2 in pig serum and named as P1 and P2.P1 and P2 factors all are circular DNA.The sizes of P1 and P2 are 648 and 993 bp,respectively.Considering the whole genome,the nucleotide homologies between P1 and PCV2,PCV1 and P2 are 32.6%-35.5%,22.9%and 63.3%. The sequence analysis reveals that the homology between the nucleotides of P1,exceptthe first16 nucleotides of 5’end,and the 107thto 739thnucleotides ofthe reverse complementof PCV2 is as high as 98.42%.However, one nucleotide of P1 that corresponds to the 461thnucleotide(A)of reverse complementof PCV2 is missing.
The harms brought by porcine circovirus-associated diseases(PCVAD)are aggravated day by day. PCV2 is immunosuppressive[11],and it generally infects animals along with other pathogens on clinic.So the early diagnosis of PCV2 infection is more important.At current,the most commonly used detection methods include ordinary PCR,immunohistochemistry and indirect immunofluorescence[12-14]. The ordinary PCR cannot analyze PCV2 quantitively.In addition,the ordinary PCR has low detection sensitivity and is susceptible to environmental pollution.The immunohistochemistry and indirectimmunofluorescence are time-consuming and operation-complicated.The real-time fluorescence quantitative PCR is a quantitative technology of nucleic acids and it is developed in recent years.Compared to ordinary PCR,the real-time PCR has higher sensitivity,specificity, speed and easier operation.It has been widely applied in genotype analysis and qualitative and quantitative detection of the viruses.The commonly used quantitative PCR methods mainly include two types,SYBR Green Ιdye and TaqMan probe.The specificity of TaqMan real-time PCR is higherthan thatofSYBR Green Ireal-time PCR.In addition,the cost of TaqMan real-time PCR is lower than that of other probe detection methods[15].
Due to the high homology between the nucleotide sequence of P1 and that of PCV2 ORF2,the detection of PCV2 with ORF2-based quantitive PCR method may be interfered by P1. Therefore,we established 2 detection methods,ORF1-based and ORF2-based.In addition,the detection results of the 2 methods were then verified with the SYBR Green I real-time PCR method established by Wen et al.[16]for PCV2-like factor P1.Our results showed the PCV2 ORF1-based detection method had higher accuracy and specificity compared to those of ORF2-based detection method,providing a rapid,sensitive and specific method for the early diagnosis and epidemiologicalinvestigation of PCV2.
Materialand Methods
Strains and infected samples
The E.coli DH5αwas purchased from the TaKaRa Biotechnology (Dalian)Co.,Ltd.The PPV,PCV1, PCV2,PRV and PRRSV were all isolated and preserved by the Key Laboratory of Veterinary Biologicals Engineering and Technology of Jiangsu Academy ofAgriculturalSciences.The 80 serum and tissue samples of pigs were randomly collected from partial regions ofJiangsu Province.
Main reagents and equipment
The DL2000 DNA Marker,pMD-18-T vector and Premix Ex TaqTM(Probe qPCR)were purchased from the TaKaRa Biotechnology(Dalian) Co.,Ltd.The 2×Taq PCR MasterMix was purchased from the Tiangen Biotech(Beijing)Co.,Ltd.The gel extraction kit and plasmid extraction kit were purchased from the AXYGEN Biotech(Hangzhou)Co.,Ltd.The Column Viral DNAout Kit and Column Animal DNAout Kit were purchased from the Tiandz Science and Technology(Beijing)Co.,Ltd.The ABI 7500 PCR System was produced by the Applied Biosystems.
Design and synthesis of primers and probes
The primers(F1,R1,F2,R2)and probes(P1,P2)for TaqMan real-time PCR were designed by using Primer Express 3.0 according to the sequences of PCV2 ORF1 and ORF2 registered in Gen Bank.The sizes of the amplified segments were all99 bp. The primers(SF1,SR1,SF2,SR2)for standard preparation were designed by using Primer Express 5.0.The sizes of the amplified segments were 945 and 702 bp,respectively.The sequences of primers and probes were shown in Table 1.All the primers and probes were synthesized by TaKaRa Biotechnology(Dalian)Co.,Ltd.
Preparation of recombinant plasmid standards
The standard preparation primers (SF1,SR1,SF2,SR2)were used to amplify PCV2.The sizes of the target genes were 945 and 702 bp,respectively.The reaction system was as follows:25.0μl 2×Taq PCR MasterMix, 1.0μlupstream primer(10μM),1.0μl downstream primer(10μM),3.0μl DNA sample,20μl sterile water.The reaction conditions were as follows: pre-denaturation at 95℃for 5 min; denaturation at94℃for 30 s,annealing at50℃for30 s(ORF2,56℃),extension at 72℃for 1 min,38 cycles; extension at72℃for 10 min.
After the amplification,the PCR products were examined by 1%agarose gel electrophoresis.Then the target fragment was recovered and then connected to pMD18-T vector forcloning.The engineering bacteria containing pMD18-T-ORF1 and pMD18-TORF2 were incubated.The plasmids were then extracted.The DNA concentration was determined with nucleic acid analyzer.The OD260nmand OD280nmwere obtained and then used to calculate the copy number of plasmid per unit volume.The plasmids were stored at-20℃.

Table 1 Names and sequences ofprimers and probes
Establishment of real-time PCR detection method
Optimization of real-time PCR reaction conditionsThe TaqMan probe method was adopted.In the ABI7500 PCR system,25μlof reaction system, 0.90-1.10 of amplification efficiency (E),>0.99 ofcorrelation coefficient,no non-specific amplification of non-template control(NTC)were treated as the standards for optimizing the concentrations of primers and probes and the annealing time and temperature.
Construction of fluorescence quantitive standard curveThe OD260nmand OD280nmof recombinant plasmids were determined.Only when the OD260nm/OD280nmranges from 1.8 to 2.0 can the recombinant plasmid be used to construct standard curve.The OD260nmwas used to calculate the copy number of plasmid.Then the plasmid was diluted into 101-1010copies/μl by 10-fold gradient with TE buffer(pH 8.0).The 10 gradients ofplasmid were used as templates.According the optimized reaction conditions,the amplification was conducted with ABI7500 PCR system.After the amplification, the standard curve was obtained automatically by using the computer-own analysis software(7500 software V2.0.5).
Sensitivity of real-time PCRThe 10-fold diluted plasmid standards were used as the templates for real-time PCR.And thus the low detection limit was determined.
Repeatability of real-time PCR
Among the 10 gradients of plasmid,3 gradients were selected.There were 3 duplications for the amplification of each gradient.After the Ct values and variation coefficientwere analyzed.
Specificity of real-time PCRThe PPV,PCV1,PRV and PRRSV were detected with the established PCR method.The PCV2-positive plasmid was used as a positive control,while the sterile water was used as a negative control.
Detection of clinicalsamples
The PCV2 in the 80 serum and tissue samples was detected by using the 2 established real-time PCR methods respectively.The detection results-inconsistent samples were then detected with the SYBR Green I realtime PCR method established for detecting PCV2-like factor P1.The 2 established methods were compared.
Results and Analysis
Preparation of plasmid standards
Identification of PCR amplification products and recombinant plasmid standardsThe genome DNA of PCV2 was extracted.It was amplified with PCV2-specific primer pairs(SF1, SR1,SF2,SR2),respectively,and then 2 band in size of945 and 702 bp were obtained(Fig.1).The bands were recovered and then connected into pMD18-T vector,respectively.The recombinant plasmids were expressed by E.coli DH5α.Then the recombinant plasmids were extracted and identified by PCR.After then,2 specific bands in size of945 and 702 bp were shown on electrophoretograms(Fig.2).The sequencing results indicated the PCR products were target fragments.And thus the plasmid standards were successfully prepared.
Concentration determination of recombinant plasmid standardsThe recombinant plasmid was diluted by 50-fold,and then the plasmid DNA concentration was determined by nucleic acid and protein analyzer.The concentrations of ORF1 and ORF2 were 3.4 and 2.5μg/ml,and the rations of OD260nmand OD280nmwere 1.95 and 1.8,indicating the purities of ORF1 and ORF2 were qualified.According to the following formula:Copy number= plasmid concentration×Avogadro constant/(Average molecular weight of 1 base pair×Totallength ofplasmid),the plasmid concentrations of ORF1 and ORF 2 could be calculated(ORF1, 4.34×1010copies/μl;ORF2,3.40×1010copies/μl).
Optimization of real-time PCR reaction conditions
The optimized reaction system (25μl)was as follows:2×Premix Ex Taq(Probe qPCR)12.5μl,upstream primer(10μM)0.5μl,downstream primer(10μM)0.5μl,ROX Reference DyeⅡ(50×)0.5μl,probe(10μM)0.5 μl,plasmid template 1.0μl,ddH2O 10.0 μl.The optimized reaction conditions were as follows:95℃30 s;95℃5 s, 57℃(ORF2,60℃)34 s,40 cycles.
Construction of real-time PCR standard curve
The 10-fold diluted gradient plasmid standards were used for real-timePCR.The dynamic amplification curves(Fig.3,Fig.4)and standard curves(Fig.5,Fig.6)of ORF1 and ORF2 were drawn automatically.The standard curves showed that there was a good linear relationship between the logarithms of initialconcentrations of plasmid templates and the Ctvalues(r2>0.99).The amplification efficiency ranged from 0.90 to 1.10. The slope of the curves ranged from -3.6 to-3.1.
Sensitivity analysis of real-time PCR
The recombinant plasmid was diluted gradiently by 10-fold until the plasmid DNA could notbe amplified by FQ-PCR,and thus the lowest copy number of template that could just be detected by FQ-PCR system is obtained.The sensitivities of ORF1 and ORF2 real-time PCR methods were 1×101(Fig.5)and 1×102copies/μlrespectively(Fig.6).The sensitivity of conventional PCR method was 1×104copies/μl.Itwas indicated the sensitivities of 2 established FQ-PCR methods were higher than that of conventional PCR by 1000(ORF1)and 100 (ORF2)times.
Repeatability of real-time PCR
The repeatability test was carried out on 1×103(S3),1×105(S5)and 1× 107(S7)copies/μlofplasmid standards of ORF1 and ORF2.The dynamic curves were shown in Fig.7(left, ORF1;right,ORF2).The results showed the variation coefficients of Ct values within groups were all lower than 5%(Table 2,Table 3).
Specificity of real-time PCR
The PPV,PCV1,PRV and PRRSV were detected with the 2 established methods respectively.Then the PCV2-positive sample was used as the positive control,while the sterile water was used as the negative control.The detection results were shown in Fig.8(left,ORF1;right,ORF2).The other viruses and negative control all showed no fluorescence signalexcept the PCV2-positive sample.It was suggested the 2 established methods all had a good specificity.
Detection of clinicalsamples
The PCV2 in the 80 collected serum and tissue samples was detected quantitively by 2 established FQ-PCR methods respectively.The results showed the detection magnitudes of 72 samples were basically consistent between the 2 detection methods,while the magnitudes of the other 8 amplified samples were inconsistent.Among the 8 samples,the detected concentrations of ORF2 were all higher than those of ORF1.So we suspected that the 8 samples mightbe infected with P1.Then the 8 samples were detected with SYBRGreen Irealtime PCR method established based on the sequence of PCV2-like factor P1 by Wen et al.The analysis on the dissolution curves showed the 8 samples were infected with P1(Table 4).
Discussion
In 1997,the post-weaning multisystemic wasting syndrome(PMWS) was firstly emerged in the SPF pigs in Canada.After then,the PMWS becomes more and more popular across the world.PCV2 often infects animals along with CSFV,PRV and PRRSV, bringing huge economic losses to pig industry.The main syndrome of PCV2 infection is to cause immune suppression,thereby leading to secondary infections and conditionalinfections.The commonly used detection method of PCV2 is conventional PCR method. But the conventional PCR method has false positivity,complex operation and low detection sensitivity.In this study, we established 2 TaqMan FQ-PCR methods.They not only could be used to detect quantitively PCV2 pathogen, but also had simple operation,high sensitivity,high specificity and good repeatability.So they can be widely used in clinicaldetection.
The currently reported real-time FQ-PCR detection methods for PCV2 mainly use the gene ofORF2 or ORF1 as the target gene to design primers and probes.It has been reported the ORF2 of PCV2 mainly encodes the capsid protein.It contains several immunoreactive sites and can react specifically with anti-PCV-2 positive sera.The ORF2 is a major immune response protein[17].Atpresent,mostof the FQ-PCRs adopt the ORF2 as the target gene.The PCV2 ORF1 encodes an essentialprotein for the duplication of viral genome.The ORF1 gene is highly conserved in all the porcine circoviruses.They are replaceable by each other in function.At current,there have also been FQPCRs using the ORF1 as the target gene.In the FQ-PCR,the effects of primers or probes will be affected by the mismatch between oligonucleotide and target sequence.This mismatch will reduce the accuracy of quantitive detection and may cause the emergence of false negativity.Therefore, the target gene of FQ-PCR should better select the highly-conserved gene fragments.The analysis on the whole genome sequence of PCV2 shows the ORF1 is more conservativethan that of ORF2 in all the sequence of PCV2.In recent years,the nucleotide sequence of P1 has been found substantially completely overlapped with that of PCV2 ORF2.And thus when the FQ-PCR uses the ORF2 as the target gene,the detection results of PCV2 may be interfered by the existence of P1 in samples. However,this interference can be completely avoided by using ORF1 as the targetgene of FQ-PCR.Therefore, the ORF1 TaqMan real-time PCR method is more specific and accurate.
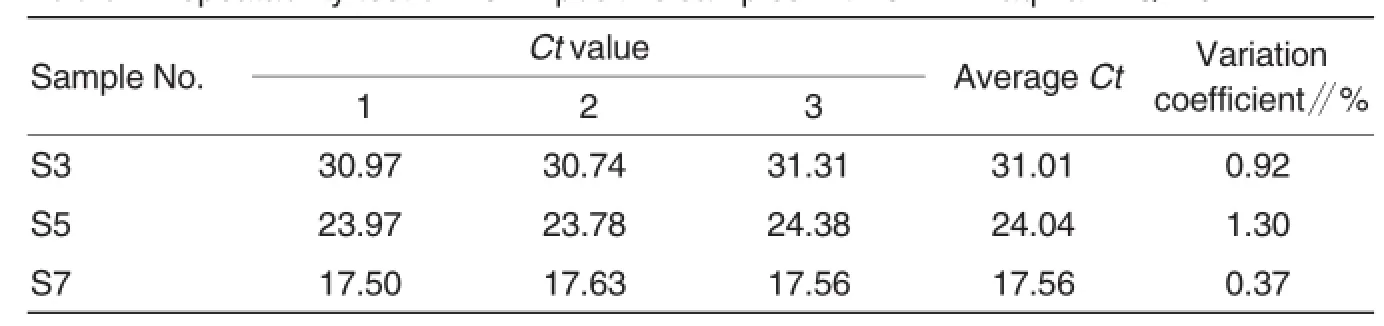
Table 2 Repeatability testofPCV2-positive samples with ORF1 TaqMan FQ-PCR
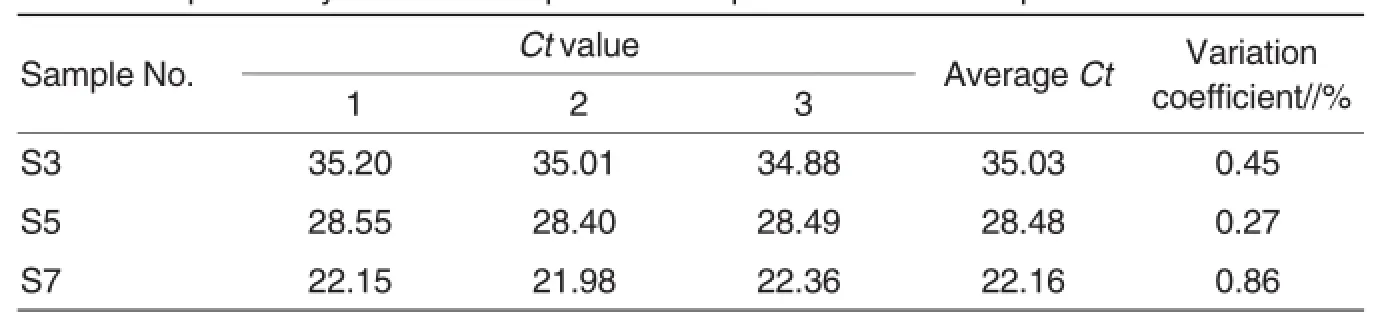
Table 3 Repeatability testof PCV2-positive samples with ORF2 TaqMan FQ-PCR
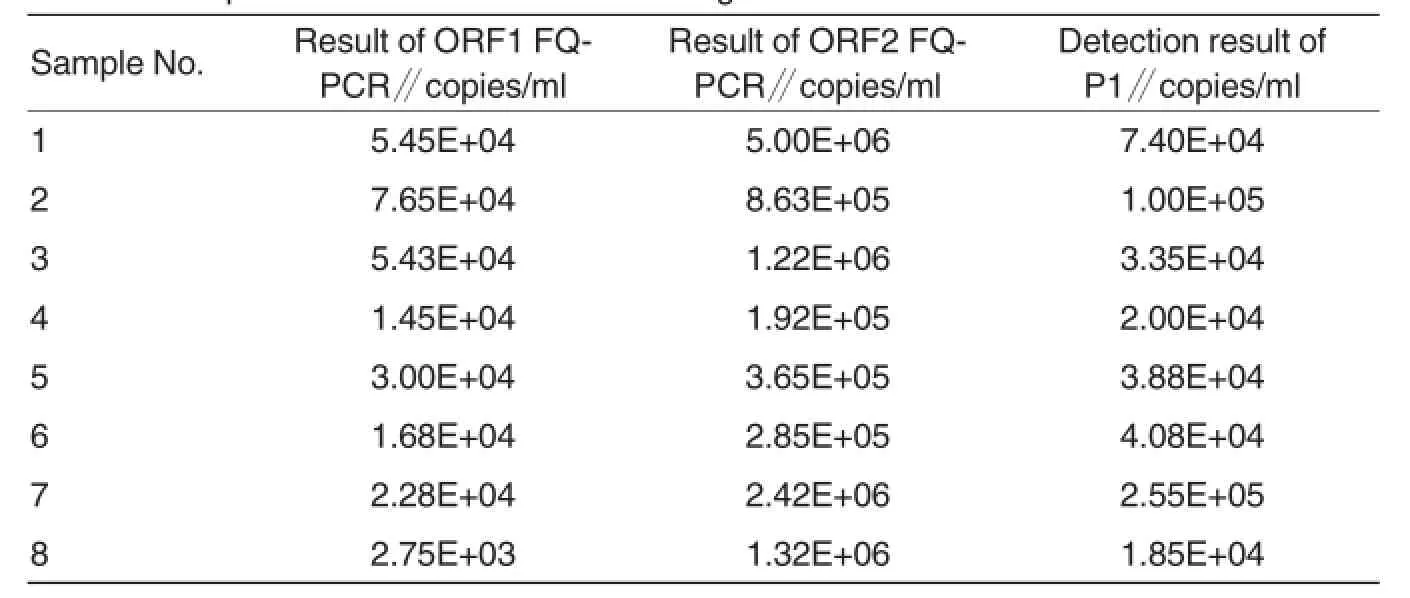
Table 4 Comparison ofdetection results among 3 FQ-PCR methods
In this study,the collected 80 samples were detected with 2 established PCV2 TaqMan FQ-PCR methods respectively.In addition,the P1 in the results-inconsistent samples were further detected with SYBR Green I FQ-PCR method.The results showed P1 existed in the results-inconsistent samples.It is suggested the quantitive detection of PCV2 may be interfered by P1 on clinic.Therefore,the PCV2 ORF1-based TaqMan FQ-PCR method has higher specificity and accuracy compared to those of PCV2 ORF2-based method.The PCV2 ORF1-based TaqMan FQ-PCR method can be used in the clinicaldetection ofPCV2.This willlay a foundation for the further researches on pathogenesis and epidemiology of PCV2.
[1]TISCHERΙ,GELDERBLOM H,VETTERMAON W,et al.Avery small porcine virus with circular single stranded DNA[J].Nature,1982,295: 64-66.
[2]ALLAN GM,MCNEILLY F,MEEHAN B, et al.Isolation and characterization of circoviruses from pigs with wasting syndromes in Spain,Denmark and Northern Ireland[J].Vet Microbiol,1999,66: 115-123.
[3]SEGALES,DOMJNGO M.Post-weaning multisystemic wasting syndrome (PMWS)in pigs[J].Veterinary Quarterly,2002,24(3):109-124.
[4]ALLAN GM,MEEHAN B,TODD D,et al.Novel porcine circovirus from pigs with wasting dis ease syndromes[J].Vet Rec,1998,142:467-468.
[5]SPILLANE P,KENNEDY S,MEEHAN B,et al.Porcine circovirus infection in the Pepublic of Ireland[J].Veterinary Record,1998,143:511-512.
[6]ELLIS,HASSARDL,CLARR E,et al. Isolation of circovirus from lesions of pigs with post-weaning multisystemic wasting syndrome[J].Can Vet,1998,39 (1):44-51.
[7]WELLENBERG G J,PESCH S,BERNDSEN FW.Isolation and characterization of Porcine circovirus type 2 from pigs showing signs of post-weaning multisystemic wasting syndrome in the Netherlands[J].Veterinary Quarterly, 2000,22(3):167-172.
[8]WEN LB(温立斌),HE KW(何孔旺), YANG HC(杨汉春).Genome sequencing and analysis of a strain of PCV2-like factor P1(一株类猪圆环病毒2型因子P1的全基因组序列测定㈦分析)[J]. Scientia Agricultura Sinica(中国农业科学),2010,43(2):411-416.
[9]WEN LB(温立斌),HE KW(何孔旺), YANG HC(杨汉春).Infectious molecular cloning of P1 factor in vitro(P1因子分子克隆的体外感染性分析)[J].Acta Veterinaria Et Zootechnica Sinica(畜牧兽医学报),2008,39(7):941-944.
[10]WEN LB(温立斌),HE KW(何孔旺), YANG HC(杨汉春),etal.Detection of cell apoptosis of immune organs induced by PCV2-like factor P1 by TUNEL method(TUNEL法检测类猪圆环病毒2型因子P1诱导猪免疫器官细胞凋亡的研究)[J].Acta Agriculturae Boreali-sinica(华北农学报),2008,23 (Plus):88-91.
[11]YANG HC(杨汉春).Epidemic features and control measures of immunosuppresive diseases ofpigs(猪免疫抑制性疾病的流行特点㈦控制对策)[J].China Animal Husbandry&Veterinary Medicine(中国畜牧兽医),2004,31(5): 41-43.
[12]LIY(李Ⅳ),YANG SX(杨世兴),WEIJZ (魏建忠),et al.Investigation on infection of PCV2 in some pig farms of AnhuiProvince(安徽省部分猪场猪圆环病毒2型感染情况调查)[J].Chinese? Journal of Preventive Veterinary Medicine(中国预防兽医学学报),2007, 29(9):732-734.
[13]HAMBERG A,RINGLER S,KRAKOWKA S.A novelmethod for the detection of porcine circovirus type 2 replicative double stranded viral DNA and nonreplicative single stranded viralDNA in tissue sections[J].J Vet Diagn Invest,2007,19(2):135-141.
[14]ALLAN GM,ELLIS JA.Porcine circoviruses:a review[J].J Vet Diagn Invest,2000,12(1):3-14.
[15]XUE S(薛霜),DU JZ(独军政),GAO SD (高闪电),et al.Research advances of real-time PCR technology and its application in veterinary medicine(实时荧光定量PCR技术研究进展及其在兽医学中的应⒚)[J].Chinese Agricultural Science Bulletin(中国农学通报),2010, 26(7):11-15.
[16]WEN LB(温立斌),HE KW(何孔旺), YANGHC(杨汉春),etal.Detection of PCV-like factor P1 with SYBR Green I real-time PCR(SYBR GreenⅠ荧光定量PCR检测类猪圆环病毒因子P1)[J]. Acta Agriculturae Boreali-sinica(华北农学报),2009,24(4):31-35.
[17]NAWAGITGUL P,HARMS PA,MOROZOV I,et al.Modified indirect porcine circovirus(PCV)type 22 based and recombinant capsid protein (ORF2)2 based enzyme 2 linked immunosorbent assays for detection of antibodies to PCV[J].Clin Diagn Lab Immunol,2002,9(1):33240.
Responsible editor:Tingting XU
Responsible proofreader:Xiaoyan WU
Copyright Authorization Statement
Should the article be accepted and published by Agricultural Science&Technology,the author hereby grants exclusively to the editorial department of Agricultural Science&Technology the digital reproduction,distribution, compilation and information network transmission rights.
周忠涛1,2,王小敏1,汪伟1,茅爱华1,温立斌1,倪艳秀1,何孔旺1*
(1.江苏省农业科学院兽医研究所,农业部兽⒚生物制品工程技术重点实验室,国家兽⒚生物制品工程技术研究中心,江苏南京210014;2.南京农业大学动物医学院,江苏南京210095)
[目的]实现在体外对猪圆环病毒2型(Porcine circovirus type 2,PCV2)的定量检测。[方法]设计合成2组特异性引物和TaqMan探针,构建分别包含PCV2 ORF1和ORF2基因全长的重组质粒作为标准品,经过反应体系和反应条件的优化,绘制标准曲线,建立2种检测PCV2的分别基于PCV2 ORF1和ORF2的Taq Man荧光定量PCR方法。[结果]所建立的2条标准曲线的Ct值㈦模板拷贝数的对数之间具有良好的线性关系,相关系数R均在0.99以上,扩增效率均在90%~110%;重复性好,组内变异系数均小于5%;特异性强,以猪细小病毒(PPV)、猪圆环病毒1型(PCV1)、猪伪狂犬病毒(PRV)、猪繁殖㈦呼吸综合征病毒(PRRSV)为模板时均无扩增;敏感性高,ORF1方法可达1.0×101copies/μl,ORF2方法可达1.0×102copies/μl。⒚建立的2种荧光定量PCR方法分别对80份临床样品进行检测并对结果进行比较,结果表明,其中72份临床样品的定量结果数量级基本一致,有8份临床样品的定量结果数量级不一致。利⒚SYBR GreenⅠ荧光定量PCR方法对8份样品进行了检测,证明这8份样品中确实存在P1的感染。[结论]基于ORF1建立的定量检测方法更为准确,可⒚于PCV2的快速诊断和定量检测。
猪圆环病毒2型;TaqMan荧光定量;ORF1;ORF2;类猪圆环病毒因子P1
国家自然科学基金(31302071);公益性行业(农业)科研专项(201303046);江苏省自主创新项目(CX(14)2045);江苏省第四期"333高层次人才培养工程"(第二层次)资助项目(BRA2012194)。
周忠涛(1986-),男,山东泰安人,硕士,主要从事人兽共患病防控研究,E-mail:zhouzhongtao08@163.com。*通讯作者,研究员,主要从事动物病毒学和细菌学研究,E-mail:kwh2003@263.net。
2014-10-17
Supported by NationalNaturalScience Foundation of China(31302071);Special Fund for Agro-scientific Research in the Public Interest(201303046);Jiangsu Agricultural Science and Technology Innovation Fund(CX(14)2045);"333 High-level Personnel Training Project"ofJiangsu Province(BRA2012194).
*Corresponding author.E-mail:kwh2003@263.net
Received:October 17,2014 Accepted:December 19,2014
修回日期2014-12-19
 Agricultural Science & Technology2015年1期
Agricultural Science & Technology2015年1期
- Agricultural Science & Technology的其它文章
- Technology Research of Plantlets Rooting and Transplanting on Tissue Culture of Ilex centrochinensis
- Analysis on Expression Patterns of NAC1 Gene in Tomato Induced by Low Temperature
- Changes of Protective Enzyme Activity and MDA Contentin Leaves of Agropyron cristatum Under Grazing Stress
- Characteristics and High-yielding Cultivation Technology of HuaimaiNo.29
- Study on Sensitivities of16 Rice Varieties to Acetochlor
- ELISA Detection of Antibody against Canine Distemper Virus in Fox
