Construction of a Full-Length cDNA Library of Solen grandis Dunker and Identification of Defense- and Immune-Related Genes
SUN Guohua,LIU Xiangquan,,REN Lihua,YANG Jianmin,WEI Xiumei,and YANG Jialong
1) Shandong Provincial Key Laboratory of Restoration for Marine Ecology,Shandong Marine Fisheries Research Institute,Yantai 264006, P.R.China
2) Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai 264003, P.R.China
1 Introduction
Solen grandisDunker,one of the most important economic marine bivalves in sands of intertidal areas at depths of 30–40 cm,widely inhabites the west coast of the Pacific Ocean from North Korea to the Philippines crossing China,Japan and the island of Timor (Poutiers,1998).In China,S.grandisis considered to be a precious marine shellfish because of its delicious taste,high nutrient content and economic value.However,overfishing and large-scale farming of other shellfish have decreased and almost exhausted the natural populations ofS.grandis(Zhanget al.,2009).As the natural resources decrease and the market demand increases,the farming industry ofS.grandisneeds further development.With improved aquaculture techniques,the basic information on the genetic characteristics ofS.grandismust be reinforced and associated important functional genes must be further investigated for the sustainable long-term development of this aquaculture industry.
Full-length cDNA libraries are indispensable tools in functional genomics and are widely used as resources to identify new genes (Wiemannet al.,2003).As genome sequencing ofS.grandisis unlikely to be accomplished in the near future,a cDNA library would be an efficient alternative for obtaining large-scale gene sequences.The construction and screening of full-length cDNA libraries have been successfully applied to aquatic animals for gene discovery.For instance,several HSP70 genes were isolated from the musselMytilus galloprovincialisby constructing a full-length cDNA library (Kourtidiset al.,2005).The immune-related genes ofEriocheir sinensiswere cloned by constructing a cDNA library of hemocytes (Zhaoet al.,2009).In addition,large quantities of expressed sequence tags (EST) from the cDNA libraries of other aquatic animals such as rainbow trout (Rexroadet al.,2003) andApostichopus japonicas(Zhouet al.,2009) have been sequenced and analyzed.
Previously,the defense mechanisms and immune systems of several shellfish species have been systematically studied (Bayne,1983; La Peyreet al.,1995; Roch,1999).However,less is known regarding the mechanisms of defense and immunity inS.grandis.To further clarify the underlying molecular mechanisms,a full-length cDNA library ofS.grandiswas constructed using the ‘switching mechanism at the 5’-end of the RNA transcript’ (SMART)technology.The resulting EST sequences were analyzed to identify the defense and immune-related genes,allowing us to study further the functional genes potentially involved in the molecular mechanisms of defense and immunity.The constructed cDNA library ofS.grandisprovided a foundation for the cloning of other functional genes as well as gene identification and utilization.
2 Materials and Methods
2.1 Sample Preparation
In November 2010,adult individuals ofS.grandis,averaging 12.5 cm in length,were collected from the sea near Rizhao,Shandong,China and held in clean sea water under laboratory conditions for two days.Tissue samples of gills and hemocytes were collected for RNA extraction and immediately preserved in liquid nitrogen.
2.2 Total RNA Extraction and mRNA Purification
Mixed tissues of gills and hemocytes were ground thoroughly in liquid nitrogen and total RNA was isolated using the Trizol reagent (Invitrogen) according to the manufacturer’s instruction.The extracted RNA was treated with DNase I to eliminate contaminant genomic DNA.The purity and integrity of RNA extract was determined using UV spectrophotometer and 1.5% denaturing agarose gel electrophoresis.The mRNA was purified using the Oligotex mRNA Kits (Qiagen).
2.3 Construction of cDNA Library
The cDNA library was constructed using the SMART cDNA Library Construction Kit (Clontech,USA) according to the manufacturer’s instruction.Poly(A)+mRNA was mixed with reverse transcription primers,i.e.,the SMART IV Oligonucleotide and the CDS III / 3’ PCR Primer,and incubated with the PowerScript Reverse Transcriptase at 42℃ to synthesize the first chain of cDNA.Then,double-stranded cDNA was obtained via long-distance PCR with the 5’ PCR Primer and the CDS III / 3’ PCR Primer with Advantage Polymerase.DscDNA was treated with proteinase K,digested with theSfiI enzyme,size fractionated,ligated with pBluescript II SK* at 16℃ and transferred intoE.coliDH-5α.The clones were stored at −80℃ with 40% glycerol.
2.4 Quality Assessment of the cDNA Library
The transformed bacteria were 10,100 and 1000 folds diluted and spread on agar plates containing IPTG (24 mg L−1)and X-gal (4 mg L−1).After 12 h incubation at 37℃,the number of clones on each plate was counted to calculate the library titer and clone capacity.The ratio of blueto-white clones was calculated to determine the recombination efficiency.To determine the size of the insert fragments,50 clones were randomly picked and cultured in 1 mL of liquid LB medium at 37℃ overnight.The target sequence was amplified using the M13 primers.
2.5 Sequence Analysis of cDNA Clones
A total of 2050 recombinants were randomly picked from the library and subjected to single-pass sequencing using the M13 primers on an automated 3730 DNA Analyzer (Applied Biosystems),of these recombinants,2038 successful yielded sequences.The sequence-documents and mask vector sequences were obtained using the software Phred (parameter-minmatch 14-penalty-2-minscore 30).The screened EST sequences were compared using the Blast software package (BlastN,BlastX),and then grouped into unigenes using the thresholds of 100 bp in length and 90% in similarity.The GoPipe package was used for statistical analysis of Gene Ontology (GO) annotations as well as gene function determination and classification.
3 Results
3.1 RNA,mRNA and cDNA Quality
The agarose gel electrophoresis clearly showed the 28S and 18S RNA bands (Fig.1).The concentration of the total RNA was 3.8 μg μL−1.The OD260/OD280ratio was 2.01.The final concentration of the purified mRNA was 50.3 ng μL−1.In total,10.1 μg total RNA was obtained,which met the required amount of cDNA library construction.The fragment size of the double-stranded cDNA ranged from 500 bp to 4000 bp,indicating that the synthesized cDNA was applicable for cDNA library construction in terms of molecular weight.

Fig.1 Electrophoresis of total RNA and cDNA of S.grandis.M,DL2000 molecular marker; a,total RNA; b,synthesized cDNA.
3.2 Quality Assessment of the cDNA Library
The titer of the constructed library was 1.05×106pfu mL−1with a capacity of 1.05×106independent clones.The recombination efficiency of the library was 98%.The approximate length of the inserted fragments varied between 800 bp to 3000 bp,and 90% clones were longer than 1000 bp (Fig.2).These results showed that a high- quality cDNA library ofS.grandiswas successfully constructed.
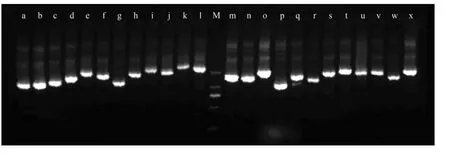
Fig.2 PCR detection of the inserts of S.grandis cDNA library.M,DL2000 molecular marker; a-x,inserts.
3.3 EST Sequence Analysis
In total,2038 ESTs (>450 bp in length) were obtained,which were clustered into 965 unigenes,of them 285 were full in length,264 were incomplete in reading frames,and 416 were undefined.GO analysis annotated only 511 of the 965 unigenes (Table 1),of them 86 were defense- and immune-related,functioning in signal transduction and communication,response to stress,and antioxidant activity.In addition,16.68% of the unigenes were involved in cell structure and processing,15.65%related to transport,metabolism and energy,and 11.72%related to replication and transcription.These unigenes were involved in 112 different GO categories including cellular component (31),molecular function (37),and biological process (44).BLASTN analysis showed that 240 of the 965 unigenes (25%) highly matched with the deposited (E-value < 1e−5; > 80% similarity) and 124 unigenes near exactly matched with the deposited (<30 bp,E-value > 1).
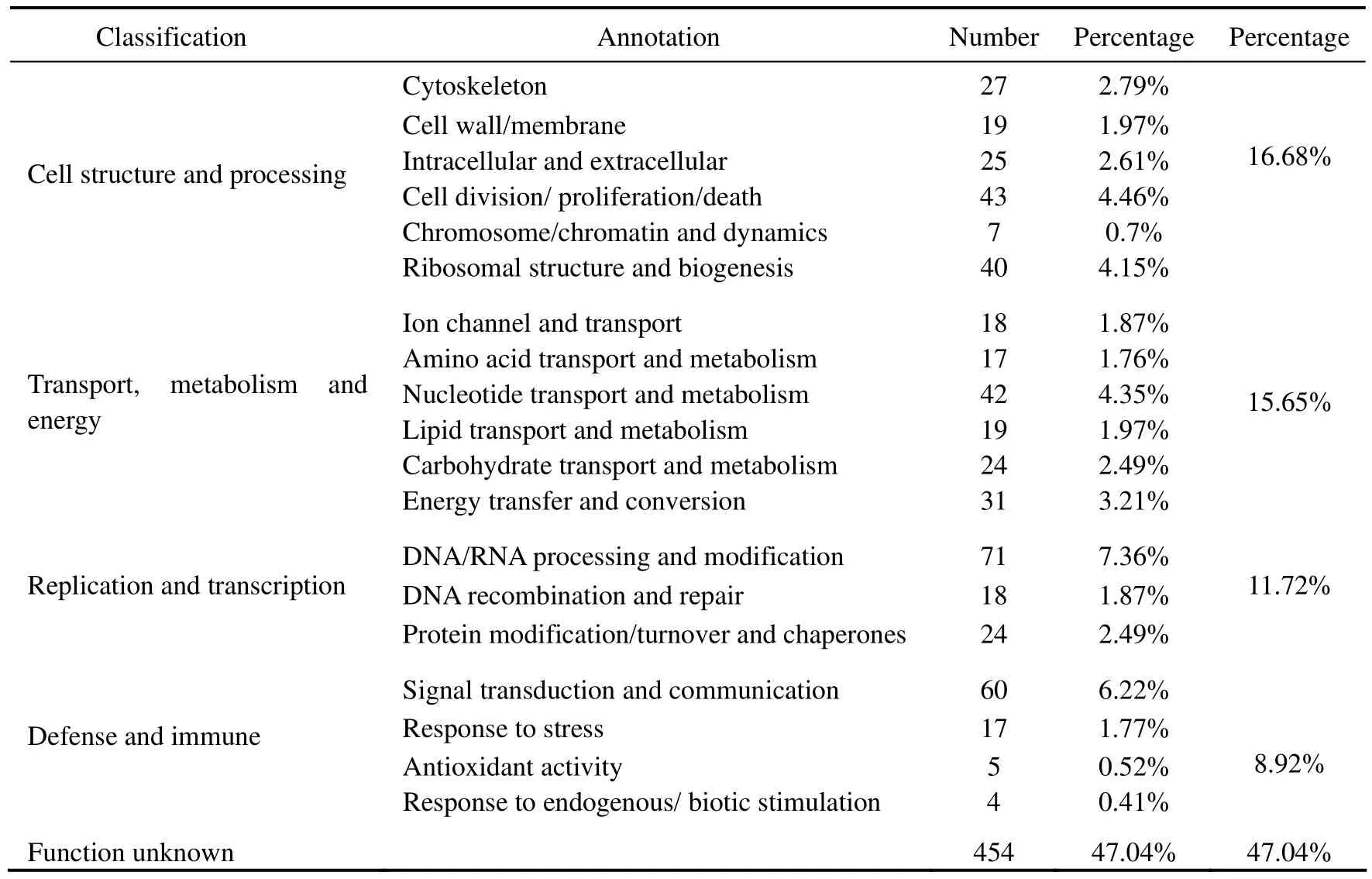
Table 1 Statistics of function and number of 965 unigenes of Solen grandis by GO analysis
3.4 Sequences of Immune-Relevant Genes
Based on GoPipe analysis and BLASTN alignments,the 15 high-identity partial sequences (E-value < 1e−10;homologous length > 80 bp) associated with defense and immunity are summarized in Table 2 and submitted to GenBank.These sequences included the stress-response proteins,e.g.,the heat-shock proteins (HSPs) and polyubiquitin,and the signaling proteins,e.g.,calmodulin,ferritin,and the guanine nucleotide-binding protein.Specifically,16 sequences were highly matched to the HSP genes,including HSP70,HSP90,and the sHSP family,whereas 11 sequences matched with the heat-shock cognate 70 (HSC70).The deduced complete 2365 bp sequence of the HSC70 cDNA contained a 5’ terminal untranslated region (UTR) of 68 bp,a 3’ UTR of 325 bp with an ‘AATAA’ tailing signal,and an open reading frame(ORF) of 1971 bp.The ORF encoded a polypeptide of 656 amino acids with an isoelectric point of 5.236 and a predicted molecular weight of 71.4 kDa.This HSC70 sequence was submitted to GenBank with the accession number DQ907944.The alignment of the complete predicted amino acid sequence with other HSC70s revealed that the HSC70 fromS.grandishad a high similarity with those from invertebrates and vertebrates.The percent similarity ofS.grandiswith that ofMeretrix meretrix,Danio rerio,andUrechis caupowas 91.2%,85.9% and 84.9%,respectively.

Table 2 High identity of partial sequences associated with defense and immune
4 Discussion
Information on the functional genes of non-model organisms is scarce.Though now RNA-seq yield large amounts of data,cDNA library construction and largescale EST sequencing have advantage in integrity and reliability of full gene sequence,and are still commonly used to isolate new functional gene sequences effectively and economically.Previous investigations onS.grandis,an economically important shellfish in the coastal areas of China,are mainly in biology,and few gene sequences ofS.grandishave been deposited in GenBank.In this study,a full-length cDNA library ofS.grandiswas constructed using the SMART technology.Approximately 2050 clones were selected for sequencing,thereby providing a full-length cDNA collection as well as other useful information resources for the isolation of the functional genes ofS.grandis.
The quality of a cDNA library can be evaluated using two critical indicators.The most important indicator is the representativeness of the library because the recombinant cDNA molecules should reflect the expression (i.e.,mRNA species) of the derived cells.This aspect is usually assayed by determination of the storage capacity of the library,which refers to the independent recombinant clones that are contained in the originally constructed cDNA library.Based on the levels of mRNA expression in eukaryotes,the storage capacity should exceed 1.7×105clones to ensure that the low-abundance cDNA would be present in the library (Sambrooket al.,2001).In the constructed cDNA library ofS.grandis,the primary library contained at least 1.05×106clones,thus was theoretically sufficient for the inclusion of rare mRNAs.The integrity of the recombinant cDNA fragments is the second indicator of the quality of a cDNA library.The SMART technology,which uses terminal transferase activity of the reverse transcriptase and the specific primers,can enrich full-length double-stranded cDNA sequences (Zhuet al.,2001).In the present study,the approximate fragment sizes of recombinant clones ranged from 800 bp to 3000 bp,which were merged into 965 unigenes.Of these,only 285 unigenes possessed complete coding frames.The constructed full-length cDNA library ofS.grandismet the requirements of a standard cDNA library.
The highly conserved protein family of HSPs is necessary to maintain protein conformation under stress conditions.It has crucial biological functions in the regulation of protein activity,heat tolerance,apoptosis,immunity,and other physiological functions (Srivastavaet al.,1994;Ishiyamaet al.,1996; Pocciaet al.,1996; Osterlohet al.,2008).In the present study,a large number of sequences in the constructed cDNA library of gills and hemocytes tissues were found related to heat shock proteins,including the HSP70 family in the majority,the HSP90 and the small HSP.The best-studied HSPs are the 70-kD protein family (HSP70) because of their role in protein chaperoning and in processes of acquired tolerance.The HSP70 family includes both heat-inducible and constitutive proteins (Ingolia and Craig,1982; Craiget al.,1983).In the present study,S.grandissamples were cultured under normal conditions without any irritation,and the obtained HSC70 sequence was presumably a constitutive isoform.It has been reported that the constitutive HSPs are expressed under normal conditions and appear to be essential for protein folding or trafficking and regulated proteolysis in unstressed cells (Craiget al.,1983; Lindquist and Craig,1988; Hightower,1993).Constitutive and inducible isoforms of the HSP70 were also reported in non-mammalian organisms such as fish (Gornatiet al.,2004; Yamashitaet al.,2004),and molluscs (Clegget al.,1998; Gourdonet al.,2000; Boutetet al.,2003).
Shellfish diseases have become more serious with the rapid advancements in aquaculture,creating a bottleneck for further development.Research on shellfish immunity is necessary to resolve these issues.Unlike mammals,aquatic organisms have rarely been studied regarding the defense systems.The identification of HSC70 and other defense genes in the present study is an effective approach towards investigation of the mechanisms of defense and immunity inS.grandis.Further research is likewise necessary to clarify the function,regulation,and induction of the spatial and temporal expression of relevant genes.
In addition to the defense- and immune-related genes,the obtained EST sequences were related to some major cellular activities and processes,including cell structure,signal transport,protein synthesis,transcription,and energy metabolism.Therefore,this work substantially contributed to the available transcriptome data ofS.grandisas well as the understanding genetic characters ofS.grandis,which would be helpful for further research on the growth,physiology,and molecular breeding ofS.grandis.In addition,this study provided information and alternative back-up technologies for protection and preservation of gene resourcesS.grandisgermplasm.
Acknowledgements
This research was supported by the Shandong Program of Agriculture Thoroughbred Project to Xiangquan Liu,the Marine Special Research Foundation of China for Nonprofit Public Industry (No.200805031),Taishan Scholar position funding of Aquatic Animal Nutrition and Feed to Linmin Zhang and NSFC funding (No.31202025).
Bayne,C.J.,1983.Molluscan Immunobiology.The Mollusca,Vol.5,Physiology,Part2.Academic Press,San Diego,407- 486.
Boutet,I.,Tanguy,A.,Rousseau,S.,Auffret,M.,and Moraga,D.,2003.Molecular identification and expression of heat shock cognate 70 (hsc70) and heat shock protein 70 (hsp70)genes in the Pacific oysterCrassostrea gigas.Cell Stress Chaperones,8: 76-85.
Clegg,J.S.,Uhlingher,K.R.,Jackson,S.A.,Cherr,G.N.,Rifkin,E.,and Friedman,C.S.,1998.Induced thermotolerance and heat shock protein-70 family in Pacific oysterCrassostrea gigas.Molecular Marine Biology and Biotechnology,7: 21-30.
Craig,E.A.,Ingolia,T.D.,and Manseau,L.J.,1983.Expression of Drosophila heat shock cognate genes during heatshock and development.Developmental Biology,99: 418-426.
Gornati,R.,Papis,E.,Rimordi,S.,Trova,G.,Sbroglia,M.,and Bernardini,G.,2004.Rearing density influences the expression of the stress-related genes in sea bass (Dicentrarchus labrax,L.).Gene,341: 111-118.
Gourdon,I.,Gricourt,L.,Kellner,K.,Roch,P.,and Escoubas,J.M.,2000.Characterization of a cDNA encoding a 72 kDa heat shock cognate protein (Hsc72) from the Pacific oyster,Crassostrea gigas.DNA Sequence,11: 265-270.
Hightower,L.E.,1993.A brief perspective on the heat-shock response and stress proteins.Marine Environmental Research,35: 79-83.
Ingolia,T.D.,and Craig,E.A.,1982.Drosophilagene related to the major heat shock-induced gene is transcribed at normal temperatures and not induced by heat shock.Proceedings of National Academy Sciences of U.S.A.,79: 525-529.
Ishiyama,T.,Koike,M.,Akimoto,Y.,Fukuchi,K.,Watanabe,K.,Yoshida,M.,Wakabayashi,Y.,and Tsuruoka,N.,1996.Heat shock-enhanced T cell apoptosis with heat shock protein70 on T cell surface in multicentric castleman’s dieease.Clinical & Experimental Immunology,106: 351-356.
Kourtidis,A.,and Drosopoulou,E.,2005.Identification of several cytoplasmic HSP70 genes from the Mediterranean mussel (Mytilusgalloprovincialis) and their long-term evolution in Mollusca and Metazoa.Journal of Molecular Evolution,62:446-459.
La Peyre,J.F.,Chu,F.L.E.,and Meyers,J.M.,1995.Haemocytic and humoral activities of eastern and Pacific oysters following challenge by the protozoanPerkinsus marinus.Fish & Shellfish Immunology,5: 179-190.
Lindquist,S.,and Craig,E.A.,1988.The heat-shock proteins.Annual review of genetics,22: 631-677.
Osterloh,A.,and Breloer,M.,2008.Heat shock proteins: linking danger and pathogen recognition.Medical microbiology and immunology,197: 1-8.
Poccia,F.,Piselli,P.,Vendetti,S.,Bach,S.,Amendola,A.,Placido,R.,and Colizzi,V.,1996.Heat-shock protein expression on the membrane of T cells undergoing apoptosis.Immunology,88: 6-12.
Poutiers,J.M.,1998.Bivalves.Acephala,Lamellibranchia,Pelecypoda.In:FAO Species Identification Guide for Fishery Purposes.The Living Marine Resources of the Western Central Pacific.Carpenter,K.E.,and Niem,V.H.,eds.,FAO,Rome,123-362.
Rexroad,III.C.E.,Lee,Y.,Keele,J.W.,Karamycheva,S.,Brown,G.,Koop,B.,Gahr,S.A.,Palti,Y.,and Quackenbush,J.,2003.Sequence analysis of a rainbow trout cDNA library and creation of a gene index.Cytogenetic and Genome Research,102: 347-54.
Roch,P.,1999.Defense mechanisms and disease prevention in farmed marine invertebrates.Aquaculture,172: 125-145.
Sambrook,T.,and Russell,D.W.,2001.Molecular Cloning: A Laboratory Manual.Vol.2,3rd edition.Cold Spring Harbor Laboratory Press,New York,313-315.
Srivastava,P.K.,and Udono,H.,1994.Heat shock proteinpeptide complexes in cancer immunotherapy.Current Opinion in Immunology,6: 728-732.
Wiemann,S.,Mehrle,A.,Bechtel,S.,WleUenreuther,R.,Pepperkok,R.,and Poustka,A.,2003.CDNAs for functional genomics and proteomics: The German consortium.Comptes Rendus Biologies,326: 1003-1009.
Yamashita,M.,Hirayoshi,K.,and Nagata,K.,2004.Characterization of multiple members of the Hsp70 family in platyfish culture cells: Molecular evolution of stress protein Hsp70 in vertebrates.Gene,336: 207-218.
Zhang,Z.W.,Yao,G.X.,and Chen,A.H.,2009.The tissue specificity and the genetic diversity of the alloyzymes ofSolen grandisDunker.Marine Sciences,33: 41-43.
Zhao,D.X.,Song,S.H.,Wang,Q.,Zhang,X.,Hu,S.,and Chen,L.,2009.Discovery of immune-related genes in Chinese mitten crab (Eriocheir sinensis) by expressed sequence tag analysis of haemocytes.Aquaculture,283: 297-303.
Zhou,Z.C.,He,C.B.,Yang,A.F.,Chen,Z.,Gao,X.G.,and Jiang,B.,2009.Construction of cDNA libraries from body wall,intestine and respiratory tree of sea cucumber (Apostichopus japonicus) and ESTs analysis.Fisheries Science,28:55-58.
Zhu,Y.Y.,Machleder,E.M.,Chenchik,A.,Li,R.,and Siebert,P.D.,2001.Reverse transcriptase template switching: A SMART approach for full-length cDNA library construction.Biotechniclues,30: 892-897.
 Journal of Ocean University of China2014年1期
Journal of Ocean University of China2014年1期
- Journal of Ocean University of China的其它文章
- Estimating the Budgets of Nutrients for Phytoplankton Bloom in the Central Yellow Sea Using a Modified Lower Tropic Ecosystem Model
- Phytoplankton Assemblage Structure Shaped by Key Environmental Variables in the Pearl River Estuary,South China
- Purification and Characterization of 2-Haloacid Dehalogenase from Marine Bacterium Paracoccus sp.DEH99,Isolated from Marine Sponge Hymeniacidon perlevis
- Molecular Phylogeny of Parapenaeopsis Alcock,1901(Decapoda: Penaeidae) Based on Chinese Materials and 16S rDNA and COI Sequence
- Cytogenetic Mechanism for the Aneuploidy and Mosaicism Found in Tetraploid Pacific Oyster Crassostrea gigas (Thunberg)
- Effect of Hydraulic Loading Rate on the Efficiency of Effluent Treatment in a Recirculating Puffer Aquaculture System Coupled with Constructed Wetlands
