Phenotypic and genotypic background underlying variations in fatty acid composition and sensory parameters in European bovine breeds
Natalia Sevane,Hubert Levéziel,Geoffrey R Nute,Carlos Sañudo,Alessio Valentini,John Williams,Susana Dunnerand the GeMQual Consortium
Background
Cattle meat provides several nutrients necessary for a balanced diet and for health preservation,especially high value proteins,minerals,B-complex vitamins and essential fatty acids(FA),and also can have an important role as a dietary source of n-3 FA and conjugated linoleic acids(CLA)[1,2].A number of epidemiological studies have associated red meat consumption with increased disease risks[3-5],whereas other authors point out the beneficial effects of the moderate consumption of unprocessed red meat-lowers total cholesterol,LDL cholesterol and triglycerides(TG)[6,7],as well as blood pressure[8].However,the isolation of the effects of red meat alone is difficult to accomplish[2].Moreover,the level of intramuscular fat content and FA composition are among the main factors determining meat palatability and consumers satisfaction[9].Muscle lipid characteristics determine meat flavour and lipid oxidation,which contributes to beef colour,and can be responsible for abnormal odours,and influences the juiciness and tenderness of meat[10].However,meat quality traits are very complex,involve many genes and are greatly influenced by a variety of environmental factors,such as diet sex,season,age,etc.[11].Being difficult and expensive to measure[12],they are usually not included in selection programs based on phenotypic performance,and challenge application of traditional selection methods,as well as the state-of-the-art Genomic Selection(GS)[13].
An alternative approach is to identify genes with an effect on fat composition and include these in selection objectives.Thanks to the genomic revolution of the past few years,more information and technology are available that can be used to improve meat quality.Many studies have identified QTL involved in meat quality related traits in beef cattle(e.g.[14,15]);however,the dissection of these QTL has not identified genetic variants explaining a large portion of phenotypic variance[16].More recently,single nucleotide polymorphisms(SNP)within candidate genes have been tested for predictive value for carcass traits,and some commercial tests to genotype animals based on SNP marker panels are being proposed to breeders(see the review in[17]).The significant progress made in characterizing changes in tissue FA composition to diet,feeding system and genotype,highlights the potential for further progress to be made through genomic or marker-assisted selection in livestock and the formulation of diets to exploit the genetic potential[18].Nevertheless,the full development of these technologies greatly depends on the precise identification of the genes and polymorphisms that have a measurable effect on muscle physiology and on meat quality,and the validation of their effects on different breeds.
In this study,we merged phenotypic and genotypic information[19-21]to characterize the variation in lipid profile and sensory parameters of Longissimus thoracis muscle and to represent the diversity among 15 cattle populations,reared under comparable management conditions.Correlations between fat content,organoleptic characteristics and lipid profiles were also investigated.
Methods
Animals and feed system
A total of 436 muscle samples from unrelated bulls belonging to 15 European cattle breeds were studied in the frame of the GeMQual(EC QLK5–CT2000-0147)European project and genotyped.The breeds included specialized beef breeds,dairy breeds,and local beef breeds.The whole sample included 31 Jersey(JER),27 South Devon(SD),30 Aberdeen Angus(AA),and 29 Highland(HIG)from United Kingdom;29 Holstein(HOL),29 Danish Red(DR),and 20 Simmental(SM),from Denmark;30 Asturiana de los Valles(AST),31 Asturiana de la Montaña(CAS),30 Avileña-Negra Ibérica(AVI),and 31 Pirenaica(PI)from Spain;30 Piedmontese(PIE),and 28 Marchigiana(MAR)from Italy;and 31 Limousin(LIM),and 30 Charolais(CHA)from France.
Bulls were reared in each country in a unique location and under a uniform beef management system representative of those used in the European Union(EU)countries.Feed composition and management details are described in[22].The diet was designed to achieve the slaughter weight of 75%of mature weight for each breed within a window of 13 to 17 mon.Animals from each breed were slaughtered the same day in either commercial or experimental abattoirs,depending on the experimental facilities of each country.
The part of this work involving live animal experimental intervention was reported by Albertí et al.[22]and followed the European standards on Care and Use of Animals(1999/575/EC).The present work was conducted on DNA and carcass samples and no Care and Use of Animals was needed.
Sampling and phenotype measurements
Carcass processing after slaughter was described by[22]and[23].For lipid measurements,Longissimus thoracis muscle was excised at 24 h postmortem from the left side of the carcass between the 6thand the 13thrib and a sample was taken immediately and frozen for chemical analysis including fat concentration.The remainder was stored at 2°C ± 1°C until 48 h post-mortem.Also,samples were taken from the 48 h post-mortem section to determine total lipid content.Samples for individual FA analysis were taken from the same position on Longissimus thoracis from all animals and vacuum packed,frozen and transported on dry ice to University of Bristol(United Kingdom)to determine total lipid content.
Fatness score(FS)corresponds to the visual fatness cover estimated by UE standard(1=very low,5=very high),and fat percentage(FP)is the proportion of subcutaneous and intermuscular fat in the rib dissection.Fat was extracted by the method of[24]separated into neutral lipid and phospholipid,methylated,separated by gas-liquid chromatography(GLC)and the individual peaks identified and quantified as described in detail by[25].Total lipid content was taken as the sum of the neutral lipid(NL)and phospholipid(PL)fractions.Some additional phenotypes were set as are saturated FA(SFA),monounsaturated FA(MUFA),polyunsaturated FA(PUFA),n-6/n-3 ratios,polyunsaturated to saturated FA(P:S)ratios and antithrombotic potential(ATT),which is the ratio between the sum of the antithrombogenic FAs eicosatrienoic acid(20:3 n-6)and eicosapentaenoic acid(20:5 n-3),and the thrombogenic FA arachidonic acid(20:4 n-6)[(20:3 n-6+20:5 n-3)/20:4 n-6][26].Sensory panel tests assessed meat using an eight-point scale as described in[27].The criteria assessed were flavour,texture and juiciness-the higher scores corresponding to the characteristic flavour of beef,and very tender and juicy meat,respectively.See[21]for detailed phenotype values per breed.
Marker selection
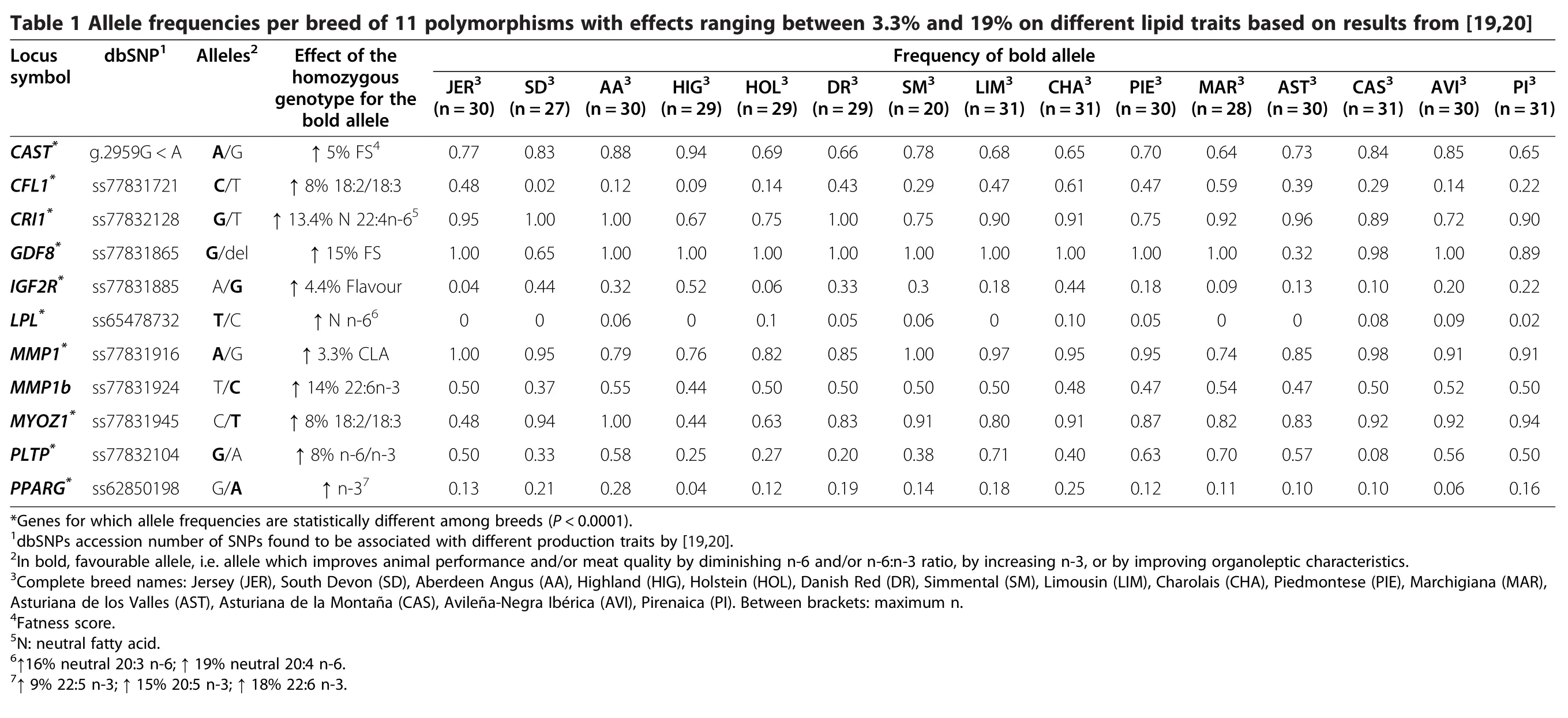
The allele frequencies of 11 polymorphisms found to be associated with different lipid traits across breeds and causing increases in traits ranging between 3.3%and 19%for one homozygous genotype compared to the other homozygous genotype(Table 1)[19,20],were used for a principal components analysis(PCA)to represent the diversity among the 15 cattle populations:calpastatin(CAST)g.2959G<A[20];cofilin 1(CFL1)ss77831721[19];EP300 interacting inhibitor of differentiation 1(CRI1)ss778332128[19];myostatin(GDF8)ss77831865[20];insulin-like growth factor 2 receptor(IGF2R)ss77831885[19];lipoprotein lipase(LPL)ss65478732[20];matrix metalloproteinase 1(MMP1)ss77831916,ss77831924[19];myozenin 1(MYOZ1)ss77831945[20];phospholipid transfer protein(PLTP)ss77832104[19];peroxisome proliferator activated receptor γ(PPARG)ss62850198[20].The six SNPs from[19]were genotyped by Kbioscience using the proprietary Kaspar©methodology;the five SNPs from[20]were genotyped by SNP multiplex and Primer Extension amplification.
Statistical analysis
Spearman correlations were determined between fatness score,fat percentage,flavour,juiciness,texture,and different lipid profiles of Longissimus thoracis muscle in 15 European cattle breeds using the CORR procedure of SAS and considering the whole set of data on all animals.Allele frequency data were subjected to ANOVA,using the PROC GLM procedure of SAS and considering breed as independent variable.A PCA procedure was performed using the mean of phenotypic measurements by breed and allelic frequencies from 11 polymorphisms to determine the main traits and SNPs that explained most of the variation among the 15 cattle populations.The frequency of the allele showing a positive correlation with the trait was used(bold allele in Table 1).All these statistical analyses were carried out using the SAS statistical package v.9.1.3[28].
Results
Trait correlations
Table 2 shows the main correlations between FS,FP,flavour,juiciness,texture,and different lipid profiles of Longissimus thoracis.FS correlated positively with FP(r=0.62,P<0.001),and both of them with absolute amounts of lipids in muscle,including total PL and NL,SFA,MUFA,PUFA,n-3 and n-6 content,as well as with flavour score.However,both of them displayed a negative correlation to P:S ratios explained by their higher correlations with SFA content(FS r=0.4,P<0.001;FP r=0.68,P<0.001)than to PUFA(FS r=0.17,P<0.001;FP r=0.44,P<0.001),and to n-6/n-3 ratios because of their lower correlations to n-6(FS r=0.13,P<0.01;FP r=0.37,P<0.001)compared to n-3(FS r=0.28,P<0.001;FP r=0.56,P<0.001).The correlation between total PL and FS(r=0.11,P<0.05)and FP(r=0.44,P<0.001)is lower than between total NL and these traits(FS r=0.44,P<0.001;FP r=0.78,P<0.001).Similar correlation coefficients were also observed between PL and total lipid(r=0.7,P<0.001)and SFA(r=0.66,P<0.001),compared to NL with both total lipid and SFA(r=0.99,P<0.001).The correlation between SFA and MUFA(r=0.99,P<0.001)was higher than between SFA and PUFA(r=0.7,P<0.001).The proportion of 18:2 n-6 declines in muscle as fat deposition increases(correlations with FP r=−0.77,P<0.001,total lipid r=−0.86,P<0.001,and SFA r=−0.88,P<0.001).
Flavour correlated positively with the organoleptic characteristics juiciness(r=0.21,P<0.001)and texture(r=0.16,P<0.001).A higher positive correlation was found between juiciness and texture(r=0.57,P<0.001).Finally,beef juiciness showed a small negative correlation to the amount of PL(r=−0.13,P<0.01)and PUFA(r=−0.1,P<0.05),particularly to n-6 content(r=−0.13,P<0.01).
Apart from results in Table 2,it is worth highlighting the positive correlation between 18:1 trans-vaccenic FA(t18:1)and CLA cis-9,trans-11(r=0.62,P<0.001).
Phenotype and genotype variation among breeds
?

Table 2 Correlations between fatness score,fat percentage,flavour,juiciness,texture,and different lipid profiles of Longissimus thoracis muscle in 15 European cattle breeds
The plot of factor pattern of the 15 cattle breeds showing the correlations to lipid traits and genotypic data from 11 polymorphisms with the two principal components is shown in Figure 1.The first two dimensions(Factor 1,42.2%;Factor 2,16.6%)explained 58.8%of the variation among breeds(Figure 1).When considering the different lipid traits,the first dimension was mainly influenced by total lipid measurements and flavour score,whereas on the opposite side muscle percentages of PUFA and of n-6,as well as P:S1,P:S2 and 18:2/18:3 ratios are plotted.The second dimension was mainly influenced by the ATT index,n-3,%n-3,and juiciness.Therefore,AA,HIG,HOL,DR and JER breeds,which displayed higher fatness[21],appeared in the positive area of the first dimension and split into two groups by the influence of the higher n-3 muscle content and flavour scores of AA and HIG breeds,and by the higher n-6 and MUFA content of HOL and DR dairy breeds.In contrast,lean breeds with high proportion of PUFAs,and high P:S and n-6 to n-3 ratios,such as PIE and AST[21],appeared at the bottom of the plot(Figure 1).Finally,SD breed stands out because of its highest ATT ratio and percentage of n-3 in muscle(Figure 1).
ConcerningSNPsdistribution,theyplottedmainly according to their previous trait associations,although also influenced by their allele frequencies per breed(Table 1).
Discussion
All animals included in this project(GeMQual QLK5–CT2000-0147)were fed a similar diet and reared intensively under comparable management conditions between countries.The effects of all factors other than breed(country,diet,slaughter)were controlled to minimize differences and were confounded with the breed effect.Inevitably,some variations might have occurred but special emphasis has been put to respect the diet composition in the different countries.The higher absolute n-3 PUFA muscle content found in the UK breeds,especially in the Aberdeen Angus breed,cannot be due to a grass-based diet generally used in UK[29]inexistent in this study,but rather to a specific characteristic of this fat breed.
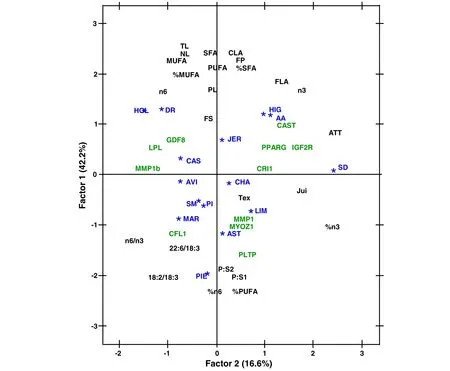
Figure 1 Plot of factor pattern for factors 1 and 2 of 15 European breeds showing the correlations to lipid traits(in black)and genotypic data(in green)from 11 polymorphisms with the two principal components.Abbreviations:TL total lipids,NL neutral lipids,PL phospholipids,FP fatness percentage,FS fatness score,Fla flavour,Tex texture,Jui juiciness,JER Jersey,SD South Devon,AA Aberdeen Angus,HIG Highland,HOL Holstein,DR Danish Red,SM Simmental,LIM Limousin,CHA Charolais,PIE Piedmontese,MAR Marchigiana,AST Asturiana de los Valles,CAS Asturiana de la Montaña,AVI Avileña-Negra Ibérica,PI Pirenaica.
Trait correlations
The correlation coefficients between total muscle lipid measurements(FS,FP,total lipids,SFA),and absolute vs.proportional(%)amounts of PUFA,n-6 and n-3 FAs were in opposite directions[30,31].For example,the sum of n-3 FA showed a positive relation to FS(r=0.28),FP(r=0.56),total lipids(r=0.63)and SFA(r=0.62)for absolute amounts,but there were negative correlations between those traits and n-3 relative proportion(FS r=−0.24,FP r=−0.34,total lipids r=−0.62,SFA r=−0.62)(Table 2).In particular,the negative correlation obtained here between the percentage of 18:2 n-6 and fat measurements,as well as the weaker correlation between total PL and FS,FP,total lipid and SFA muscle content compared with NL(Table 2)[32],is in accordance with the expected proportions of PL vs.NL as animal fattens.Long chain PUFAs are mainly stored in muscle PL in cattle,which is an essential component of cell membranes and its amount remains fairly constant as the animal fattens,whereas NL increases in overall FA composition.SFA and MUFA are mainly stored in the NL fraction in triglycerides.This means increasing carcass fat leads to an increasing amount of FAs in triglycerides,but at the same time the relative amount of PUFAs is decreasing[32],which is in concordance with the negative correlation obtained here between the percentage of PUFA and fat measurements,as well as the weaker correlation between total PL and total lipid(r=0.7)muscle content compared with NL(r=0.99)(Table 2).
In agreement also with previous studies[33,34],we found a positive correlation between t18:1 and CLA(r=0.62,P<0.001),explained by the metabolic relationships between both FA–in ruminants the SCD enzyme forms also CLA from t18:1 in adipose tissue[29,32].
Although high levels of long chain n-3 PUFA have been described as having an impact on flavour to produce a ‘grass fed’taste[35,36],and[37]found no correlation between n-6 PUFA and flavour in two beef breeds,here there was only a negative correlation to the percentage of total PUFA,particularly to n-6 fraction,whereas the percentage of n-3 in muscle did not seem to influence meat flavour in cattle not fed with a grassbased diet.
As expected,the correlation between juiciness and texture is higher than with flavour scores given that juiciness depends mainly on the meat water-binding capacity and plays a key role in meat texture[38-40],contributing to its variability[41],whereas flavour is mainly influenced by FA composition and marbling[42],as reflected by its positive correlations with all absolute fat content measurements obtained here(Table 2).Although texture and juiciness properties also are dependent on other characteristics of meat,including fat content[40],both of them showed few or no correlation with muscle fat content or FA profile(Table 2).
Phenotype and genotype variation among breeds
The distribution of breeds plotted by PCA analysis fell into three main groups(Figure 1):one group defined as having a high absolute fat content,which splits into two blocks–AA and HIG breeds on one hand,characterized by higher n-3 muscle content and flavour scores,and in the other hand HOL and DR dairy breeds,which displayed higher values of MUFA and n-6-;a second group with lower fat content and higher proportion of PUFAs,as well as PUFA vs.SFA ratios(healthier meat)PIE and AST;and a large group gathering the rest of breeds with intermediate fat content,among which it is worth highlighting SD because of its highest ATT ratio(index higher values better for health)and percentage of n-3 in muscle.
Regarding SNPs distribution,most of them plotted according to their previous trait associations and also influenced by their allele frequencies per breed(Table 1,Figure 1):PPARG,which influences the amount of 22:5 n-3,20:5 n-3 and 22:6 n-3 in muscle[20],appeared near n-3 and ATT ratio-calculated as(20:3 n-6+20:5 n-3)/20:4 n-6-factor patterns,as well as almost equidistant to the three breeds with higher allele frequencies for the A allele–AA,CHA and SD-;CFL1,PLTP and MYOZ1,which were associated with n-6 to n-3 ratio[19],correlated with 18:2 n-6/18:3 n-3 and specially with n-6/n-3 ratio in Factor 1;IGF2R,previously linked to an increase in flavour[19],shared Factor 2 pattern with flavour and was placed almost equidistant to the three breeds with higher allele frequencies for the G allele–HIG,CHA and SD-;CAST was associated with an increase in FS and appeared closely related to the two breeds with higher allele frequencies for the A allele–AA and HIG,sharing also Factor 1 pattern with FS;LPL,associated with the increase of several neutral n-6[20],plotted in the same Factor 2 pattern than n-6 content,but closer to and almost equidistant from the three breeds with higher allele frequencies for the T allele–HOL,AVI and CAS;and,as expected,GDF8 SNP was placed near the trait FS[20].
Finally,there were no relationships between the two SNPs in the MMP1 gene and the SNP in CRI1 neither with their main trait associations–CLA,22:6 n-3 and 22:4 n-6,respectively[19],nor with breed allele frequencies,which may be caused by the other trait associations of this SNPs with lower or unknown effects[19].
Conclusions
The wide range of traits and breeds studied,along with the genotypic information on polymorphisms previously associated with different lipid traits,provide a broad characterization of the phenotypic and genotypic background underlying variations in FA composition and sensory parameters between breeds,which allows giving a better response to the variety of consumers’preferences.Also,the development and implementation of low-density SNP panels with predictive value for economically important traits,such as those summarized here,may be used to improve production efficiency and meat quality in the beef industry as a molecular signature of GTTdelGCA CCAA for CAST(g.2959G<A),CFL1(ss77831721),CRI1(ss77832128),GDF8(ss77831865),IGF2R(ss778 31885),LPL(ss65478732),MMP1(ss77831916,ss77831 924),MYOZ1(ss77831945),PLTP(ss77832104),and PPARG(ss62850198),respectively,which would correspond to the “most favourable”haplotype.
Competing interests
The authors declare that they have no competing interests.
Authors’contributions
NS carried out the molecular genetic studies,performed the statistical analysis and drafted the manuscript.SD,HL,GN,CS,AV and JLW conceived
the study,and participated in its design and coordination.SD also helped to draft the manuscript.All authors read and approved the final manuscript.
Acknowledgements
This work was supported by an EC grant QLK5–CT2000-0147.
GeMQual Consortium:Albertí P Centro de Investigación y Tecnología Agroalimentaria,Gobierno de Aragón,50080,Zaragoza,Spain;Amarger V.
Delourme D.Levéziel H.INRA,UMR 1061,87000 Limoges,France and Université de Limoges,UMR 1061,87000 Limoges;Boitard S.Mangin B.INRA Chemin de Borde-Rouge-Auzeville,BP 52627,31326 Castanet-Tolosan cedex,France;Cañón J.Checa ML.Dunner S.García D.Miranda ME.Pérez R.Dpto de Producción Animal,Facultad de Veterinaria,28040 Madrid,Spain;Christensen M.Ertbjerg P.Department of Food Science,University of Copenhagen,1958 Frederiksberg C.,Denmark;Crisá A.Marchitelli C.Valentini A.Dipartimento di Produzioni Animali,Università della Tuscia,via De Lellis,01100 Viterbo,Italy;Failla S.Gigli S.CRA,Istituto Sperimentale per la Zootecnia,00016 Monterotondo,Italy;Hocquette JF.INRA,UR1213,Unité de Recherches sur les Herbivores,Centre de Clermont-Ferrand./Theix F-63122,France;Nute G.,Richardson I.Division of Farm Animal Science,University of Bristol,BS40 5DU,United Kingdom;Olleta JL.,Panea B.,Sañudo C.Dept de Producción Animal y Ciencia de los Alimentos,Universidad de Zaragoza,50013,Zaragoza,Spain;Razzaq N.Roslin Institute,Roslin,Midlothian,Scotland.EH25 9PS,UK;Renand G.INRA,UR337,Station de Génétique Quantitative et Appliquée,78352 Jouy-en-Josas cedex,France;Williams.JL.Parco Tecnologico Padano,Via Einstein,Polo Universitario,26900 Lodi,Italy.
Author details
1Departamento de Producción Animal,Facultad de Veterinaria,Universidad Complutense de Madrid,Madrid,Spain.2INRA,UMR 1061,F-87000 Limoges,France.3Université de Limoges,UMR 1061,F-87000 Limoges,France.4Division of Farm Animal Science,University of Bristol,Bristol BS40 5DU,UK.5Departimento de Producción Animal y Ciencia de los Alimentos,Universidad de Zaragoza,50013 Zaragoza,Spain.6Dipartimento di Produzioni Animali,Università della Tuscia,via De Lellis,01100 Viterbo,Italy.7Parco Tecnologico Padano,Via Einstein,Polo Universitario,26900 Lodi,Italy.
Published:15 April 2014
1.Givens DI,Gibbs RA:Current intakes of EPA and DHA in European populations and the potential of animal-derived foods to increase them.P Nutr Soc2008,67:273–280.
2.McAfee AJ,McSorley EM,Cuskelly GJ,Moss BW,Wallace JM,Bonham MP,Fearon AM:Red meat consumption:an overview of the risks and benefits.Meat Sci2010,84:1–13.
3.Cross AJ,Leitzmann MF,Gail MH,Hollenbeck AR,Schatzkin A,Sinha R:A prospective study of red and processed meat intake in relation to cancer risk.PLos Med1973,2007:4.
4.World Cancer Research Fund/American Institute for Cancer Research:Food,nutrition and the prevention of cancer:A global perspective.Washington DC:American Institute for Cancer Research;2007.
5.Kontogianni MD,Panagiotakos DB,Pitsavos C,Chrysohoou C,Stefanadis C:Relationship between meat intake and the development of acute coronary syndromes:The CARDIO2000 case–control study.Eur J Clin Nutr2008,62:171–177.
6.Beauchesne-Rondeau E,Gascon A,Bergeron J,Jacques H:Plasma lipids and lipoproteins in hypercholesterolaemic men fed a lipid-lowering diet containing lean beef,lean fish,or poultry.Am J Clin Nutr2003,77:587–593.
7.Bradlee ML,Singer MR,Moore LL:Lean red meat consumption and lipid profiles in adolescent girls.J Hum Nutr Diet2013,2:292–300.
8.Hodgson J,Burke V,Beilin LJ,Puddey IB:Partial substitution of carbohydrate intake with protein intake from lean red meat lowers blood pressure in hypertensive persons.Am J Clin Nutr2006,83:780–787.
9.Lee SH,Park EW,Cho YM,Kim SK,Lee JH,Jeon JT,Lee CS,Im SK,Oh SJ,Thompson JM,Yoon D:Identification of differentially expressed genes related to intramuscular fat development in the early and late fattening stages of hanwoo steers.J Biochem Mol Biol2007,40:757–764.
10.Bernard C,Cassar-Malek I,Le Cunff M,Dubroeucq H,Renand G,Hocquette JF:New indicators of beef sensory quality revealed by expression of specific genes.J Agr Food Chem2007,55:5229–5237.
11.Hocquette JF,Botreau R,Picard B,Jacquet A,Pethick DW,Scollan ND:Opportunities for predicting and manipulating beef quality.Meat Sci2012,92:197–209.
12.Simm G,Lambe N,Bünger L,Navajas E,Roehe R:Use of meat quality information in breeding programmes.InImproving the sensory and nutritional quality of fresh meat.Edited by Kerry JP,Ledward D.UK:Woodhead Publishing Ltd;2009:680.
13.Luan T,Woolliams JA,Lien S,Kent M,Svendsen M,Meuwissen TH:The accuracy of Genomic Selection in Norwegian red cattle assessed by cross-validation.Genetics2009,183:1119–1126.
14.Casas E,Shackelford SD,Keele JW,Stone RT,Kappes SM,Koohmaraie M:QTL affecting growth and carcass composition of cattle segregating alternate forms of myostatin.J Anim Sci2000,78:560–569.
15.Casas E,Shackelford SD,Keele JW,Koohmaraie M,Smith TPL,Stone RT:Detection of quantitative trait loci for growth and carcass composition in cattle.J Anim Sci2003,81:2976–2983.
16.Van Eenennaam AL,Li J,Thallman RM,Quaas RL,Dikeman ME,Gill CA,Franke DE,Thomas MG:Validation of commercial DNA tests for quantitative beef quality traits.J Anim Sci2007,85:891–900.
17.Ibeagha-Awemu EM,Kgwatalala P,Zhao X:A critical analysis of production-associated DNA polymorphisms in the genes of cattle,goat,sheep,and pig.Mamm Genome2008,19:591–617.
18.Shingfield KJ,Bonnet M,Scollan ND:Recent developments in altering the fatty acid composition of ruminant-derived foods.Animal2013,7:132–162.
19.Dunner S,Sevane N,García D,Levéziel H,Williams JL,Mangin B,Valentini A,GeMQual Consortium:Genes involved in muscle lipid composition in 15 European Bos taurus breeds.Anim Genet2013,44:493–501.
20.Sevane N,Armstrong E,Cortés O,Wiener P,Pong Wong R,Dunner S,GeMQual Consortium:Association of bovine meat quality traits with genes included in the PPARG and PPARGC1A networks.Meat Sci2013,94:328–335.
21.Sevane N,Cañón J,Dunner S:GemQual Consortium:Muscle lipid composition in bulls from fifteen European breeds.Livestock Sci2014,160:1–11.
22.Albertí P,Panea B,Sañudo C,Olleta JL,Ripoll G,Ertbjerg P,Christensen M,Gigli S,Failla S,Concetti S,Hocquette JF,Jailler R,Rudel S,Renand G,Nute GR,Richardson RI,Williams JL:Live weight,body size and carcass characteristics of young bulls of fifteen European breeds.Livest Sci2008,114:19–30.
23.Christensen M,Ertbjerg P,Failla S,Sañudo C,Richardson RI,Nute GR,Olleta JL,Panea B,Albertí P,Juárez M,Hocquette JF,Williams JL:Relationship between collagen characteristics,lipid content and raw and cooked texture of meat from young bulls of fifteen European breeds.Meat Sci2011,87:61–65.
24.Folch J,Lees M,Stanley GHS:A simple method for the isolation and purification of lipids from animal tissues.J Biol Chem1957,226:497–509.
25.Scollan ND,Choi NJ,Kurt E,Fisher AV,Enser M,Wood JD:Manipulating the fatty acid composition of muscle and adipose tissue in beef cattle.Br J Nutr2001,85:115–124.
26.Ulbricht TLV,Southgate DAT:Coronary heart disease:seven dietary factors.Lancet1991,338:985–992.
27.Wood JD,Nute GR,Fursey GAJ,Cuthbertson A:The effect of cooking conditions on the eating quality of pork.Meat Sci1995,40:127–135.
28.SAS Institute Inc:Statistical Analysis with SAS/STAT®Software V9.1.3.Cary,NC,USA:SAS Institute Inc;2009.
29.Scollan ND,Hocquette JF,Nuernberg K,Dannenberger D,Richardson RI,Maloney A:Innovations in beef production systems that enhance the nutritional and health value of beef lipids and their relationship with meat quality.Meat Sci2006,74:17–33.
30.Dinh TTN:Lipid and Cholesterol Composition of the Longissimus Muscle from Angus,Brahman,and Romosinuano.InMaster of Science Thesis in Animal and Food Science.Graduate Faculty of Texas Tech University:Dinh TTN;2007.
31.Hoehne A,Nuernberg G,Kuehn C,Nuernberg K:Relationships between intramuscular fat content,selected carcass traits,and fatty acid profile in bulls using a F2-population.Meat Sci2012,90:629–635.
32.Wood JD,Enser M,Fisher AV,Nute GR,Sheard PR,Richardson RI,Hughes SI,Whittington FM:Fat deposition,fatty acid composition and meat quality:A review.Meat Sci2008,78:343–358.
33.Enser M,Scollan N,Choi N,Kurt E,Hallett K,Wood J:Effect of dietary lipid on the content of CLA in beef cattle.Anim Sci1999,69:143–146.
34.Lawless F,Stanton C,L’Escop P,Devery R,Dillon P,Murphy JJ:Influence of breed on bovine milk cis-9,trans-11-conjugated linoleic acid.Livest Prod Sci1999,62:43–49.
35.Wood JD,Richardson RI,Nute GR,Fisher AV,Campo MM,Kasapidou E,Sheard PR,Enser M:Effects of fatty acids on meat quality:a review.Meat Sci2004,66:21–32.
36.Warren HE,Scollan ND,Nute GR,Hughes SI,Wood JD,Richardson RI:Effects of breed and a concentrate or grass silage diet on beef quality in cattle of 3 ages.II:Meat stability and flavour.Meat Sci2008,78:270–278.
37.Costa P,Lemos JP,Lopes PA,Alfaia CM,Costa AS,Bessa RJ,Prates JA:Effect of low-and high-forage diets on meat quality and fatty acid composition of Alentejana and Barrosã beef breeds.Animal2012,6:1187–1197.
38.Harries JM,Rhodes DN,Chrystall BB:Meat texture:I.Subjective assessment of the texture of cooked beef.J Texture Stud1972,3:101–114.
39.Dransfield E,Francombe MA,Whelehan OP:Relationships between sensory attributes in cooked meat.J Texture Stud1984,15:33–48.
40.Dransfield E,Nute G,Roberts T,Boccard R,Touraille C,Buchter L,Casteels M,Cosentino E,Hood D,Joseph R:Beef quality assessed at European research centres.Meat Sci1984,10:1–20.
41.Juarez M,Aldai N,Lopez-Campos O,Dugan MER,Uttaro B,Aalhus JL:Beef Texture and Juiciness.InHandbook of Meat and Meat Processing.Edited by Hui YH.Boca Raton:CRC press;2011:177–206.
42.Melton SL,Amiri M,Davis GW,Backus WR:Flavor and chemical characteristics of ground beef from grass-,forage-,grain-and grain-finished steers.J Anim Sci1982,55:77–87.
 Journal of Animal Science and Biotechnology2014年2期
Journal of Animal Science and Biotechnology2014年2期
- Journal of Animal Science and Biotechnology的其它文章
- Utilization of tea grounds as feedstuff for ruminant
- A note on the consistency of a behavioral play marker in piglets
- Effects of inorganic or organic selenium on immunoglobulins in swine
- Chicory(Cichorium intybus L.)and cereals differently affect gut development in broiler chickens and young pigs
- Treatment of wheat straw using tannase and white-rot fungus to improve feed utilization by ruminants
- Pigs fed camelina meal increase hepatic gene expression of cytochrome 8b1,aldehyde dehydrogenase,and thiosulfate transferase
