Isolation and Characterization of a Serial of Novel Fostriecin Analogues from FosJ-Deletion Mutant of Streptomyces pulveraceus Suggesting a Novel Polyketide Biosynthesis Insight
KONG Ri-xiang,MA Chun-yan,TANG Li
Research Center for Molecular Medicine,Dalian University of Technology,Dalian 116024,China
Introduction
Polyketiedes made by polyketide synthases(PKSs)consist of a large family of medicinally important natural products with diverse structures and biological activities[1].Depending on the nature of their constituent catalytic domains,these megasynthases generate chemical variety and complexity in a stepwise fashion[2].In general,modular PKS use an assembly line mechanism,and most modules follow rules that a typical module selects a starter(including acetyl-CoA or propionyl-CoA)or extender(such as a malonyl-CoA.,methylmalonyl-CoA or ethylmalonyl-CoA)unit based on its specificity of a unique acyl transferase(AT)domain and condenses wiyth the growing chain using its β-ketoacyl synthase(KS)domain.An acyl carrier protein(ACP)domain acquires the extender polyketiede chain,which can be further modiefied at the nascentβketone by the action ofβ-ketoacyl reducatase(KR),dehyrdase(DH)or enoyl reductase(ER)domains.The growing chain is then transferred from the ACP to a downstream KS,and the further rounds of elongation and processing continue until the full-length polyketide chain is completed which is typically released from the PKS by a terminal thioesterase(TE)to form a macrocyclic lactone ring or a linear product.
Fostriecin(CI-920)originally isolated from the fermentation culture of Streptomyces pulveraceus,is a clas-sic of structurally novel phosphate ester natural product which structurally related to cytostatin,leptomycin,phoslactomycin and others[3,4].It exhibits a variety of biological properties such as a cell cycles inhibitor which mostly derives from the inhibition of topoisomerase typeⅡ(TopoⅡ)or a selective inhibition of protein phosphorylation with an excellent 104fold specific-selectivity for PP2A/PP4 versus PP1,which plays an important role in the signal transduction,growth and cell division[5-8].The comprehensive biosynthesis mechanism of fostriecin has been investigated and confirmed by gene knock-out and compound isolation method[9,10].As a polykeide antibiotic,its biosynthesis is obeyed by the strict polyketide biosynthesis fashion or routine as general polyketide antibiotic.In fostriecin biosynthesis,a classic hydroxyl group is shaped at the C-3 during PKS elongation.Then,the nascent polyketide chain is circled to afford compound 4 with a six-membered ring and a malonated moiety adheres to C-3 hydroxyl group by TE and an unknown enzyme simultaneously and respectively[9].From then on,a serial of post-PKS tailoring enzymes begin to perform their functions to produce fostriecin.

Fig.1 The structure of fostriecin and its analogues
In order to investigate the biosynthesis mechanism of fostriecin and find more novel potential candidates or analogues,fosJ gene-deletion mutant was fermentated.Five minor and novel analogues(10-14)were isolated and characterized accompanied with the normal fostriecin intermediates accumulations(Figure1).The detailed structure data was listed in Table 1-3.All compounds except for 10 were the general polyketide chain without six-membered ring as fostriecin.Based on the general of polyketide biosynthesis theory,the formation of C2-C3 saturated bond should be derived from the joint function of different polyketide synthases including ketoreductase(KS),ketoreductase(KR),dehydratase(DH),and enoyl reductase(ER)in PKSs.However,DH and ER domains were not found in module 9 except for one solo set of DH/ER domain existing in module 1 in fostriecin PKSs.The C3-OH was used to create a double-bond during the post-PKS modification which has been confirmed in the previous work[9,10].Hence,the saturated bond at C2-C3 must come from the other potential reaction or a potential enzyme functionswhich maybe contrary againstthe common polyketide biosynthesis.It will be an interesting phenomenon and bring a novel insight to elucidate the mechanism polyketide biosynthesis.Here,the detailed structure was reported and some possible biosynthesis pathways of these compounds were proposed(Figure 2).
Materials and Methods
General experimental procedures
The NMR spectra were recorded on Bruker Avance II400 spectrometer(400 MHz for1H and 100.61 MHz for13C,Brukere,Faellanden,Switzerland)with tetramethylsilane(TMS)as an internal reference.The QTOF-HRMS and API-ES-MS data were obtained on a UPLC/Q-TOF micro mass spectrometer and HP1100MSD.HPLC analysis was performed on Agilent 1200 series with DAD detector;the sample for NMR analysis was purified with the semi-preparative column(ZORBAX SB-C18,5 um,9.4 mm × 250 mm).The general chromatography was performed on C18(40 um,YMC,Co.,Ltd.,Kyoto,Japan)or Diaion HP20SS(Mitsubishi Chemical Co.,Japan).
Inoculation preparation
The fosJ gene-deletion mutant from Streptomyces pulveraceus ATCC 31906 was constructed and confirmed as STQ0701 in our laboratory.50 milliliter of frozen cells of fosJ deletion mutant maintained in 20%glycerol was inoculated into 50 mL of Yeast Peptone Dextrose medium(YPD)in a 250 mL unbaffled flask.The primary seed culture was incubated at 28℃and 220 rpm on a rotary shaker(E25R,NBS,USA)for 24 hours.Secondary seed cultures were generated by transferring 10 mL of the primary seed culture into 500 mL YPD medium in 2 L flask.They were grown at same conditions for 1 day and used to inoculate the production cultures.
Production of 10-14
The compounds 10-14 were produced as the by-products of 3,4 and 5[9].Five-liter bioreactors(B-DCU5,B.Bruns,Germany)each containing 4 L of fermentation medium consisting of 5%glycerol,0.4%baker yeast,0.5%Meat extract,0.1%NaCl,0.25%Ca-CO3,0.25%K2HPO4,and 0.5%grains distillers(the pH value was adjusted to 7.0)were autoclaved at 121℃for 60 minutes and inoculated with 200 mL secondary seed cultures.The fermentations were performed at 28℃for 4 days with an aeration rate of 0.5 v/v/m.Dissolved oxygen was controlled at 20%of air saturation by an agitation cascade.Foaming was controlled by the addition of antifoam agent.Culture pH was monitored and controlled at 7.0 with 1mol/L NaOH or not controlled respectively.
Isolation and purification of 10-14
The afforded fermentation broth was centrifuged to remove the mycelia and the supernatant was loaded on the HP20SS equilibriumed with 0.05 M phosphate buffer(pH 6.8),then the column was eluted with 100%methanol,the fractions containing fostriecin analogues were collected,and evaporated to afford an oily residue.The oily residue was dissolved in CH3OH and diluted to 10%,then loaded on C18chromatography with the methanol-buffer as mobile phase.A stepped gradient elution of methanol-buffer(10% ,20% ,30% ,40% ,50% ,60%methanol in buffer)was used to eluted the column.All fractions containing fostreicin analogues were concentrated in vacuo and exchanged with HP20SS column.The exchanged methanol solvent was then evaporated under reduced pressure to produce an oily residue.
For compound purification,the samples were subjected to a final purification by semi preparative chromatography(ZORBAX SB-C18,250 mm × 9.4 mm 5 μm)to give pure objective compounds.
The isolated compounds were subjected structure determination with HR-MS and NMR spectrum analysis as shown at Table1,Table 2 and Table3.
Results and Discussions
Production and Isolation of 10-14
When fosJ gene-deletion mutant of Streptomyces pulveraceus was fermentated with pH value not controlled,compound 3,4,5 were accumulated as major metabolites[9],accompanied with a serial minor of metabolites isolated as compound 10-14.In order to achieve more information and further demonstrate the biosynthesis mechanism of fostriecin,they were collected,isolated and structurally characterized.However,an interesting phenomenon was observed that when the pH value was controlled at 7.0 during the whole culture process,compound 11 was produced as the prominent metabolite accompanied with a minor amount of 4,5 and 10,11-14.Two different fermentation conditions led to different productivity of their metabolites made the research more useful and interesting.
Structure elucidation
The characteristic triple-peak absorbance and 267 nm maxima in the UV spectrum of all isolated analogues,exhibited the presence of conjugated triene fragment.All of proton and carbon signals of C12-C17 in their NMR analysis were keeping consistent with forstriecin or other intermediates.All these indicated they belonged to the same structure analogues.
Compound 10 was obtained as a light-yellow oil,and its molecular formula was determined to be C22H32O9by Q-TOF-HRMS at m/z 439.3080[M-H](calculated as 439.3078),18 mass units more than of 4,consistent with a H2O mass weight.So it may be a hydrolysis product of 4.Compared to1H NMR spectrum of 4,the proton signal of C-5 of 10 disappear instead of an obvious signals was observed at δH3.97(1H,m)indicating the hydration occurs at C1-C5 position.A triplet signal of δH3.20(2H,t,4)at C-2'and δH2.55(1H,dd,14.4,6.5)/2.40(1H,dd,14.5,7.0)proton signal deriving from the coupling interactions of the proton in the C-3 position and other nonequivalent protons indicated a malonic-acid moiety existing in it structure as 4.To the mentioned that an H-D exchange phenomenon did not occurs during the NMR analysis of 10 like 4.In the13C NMR spectrum,the carbon signal of C-5 was upfield shifted to δc71.0,the C-2 and C-3 signal was downfield shifted to δc41.6 and δc41.3 whereas the δc48.4 signal of C2'-CH2was discerned easily.Thus,the structure of 10 was characterized as a new fostriecin analogue.
Q-TOF-HRMS of 11 revealed a molecular formula of C19H30O5at m/z 337.2012[M-H]-(calculated as 337.2015),16 mass units less than that of 15(it could not be achieved to the date),and consistent with a dehydroxy analog of 15.The1H NMR spectrum of 11 revealed the absence of the C2'and C3'1H resonance at δH7.02 and δH5.97 ordinarily seen in fostriecin,instead of δH2.07(2H,t)and δH1.29-1.60(2H,m).Moreover,the 8-CH3proton signal being upfield shifted to δH0.94(2H,d,6.8)from δH1.17(3H,s)in 10 and an obvious δH2.11(1H,m)proton signal is observed in 11,suggests the absence of C8-OH.Moreover,two downfield carbon signal at δC120 and δC146 in13C NMR of fostriecin was exchanged with two high field carbon signal at δC38.5 and δC23.7,indicating the C2-C3 double bond was reducted to a saturated bond.Accordingly,the formation of saturated bond leads to absordance decreasement at 220 nm in the UV spectrum of 11.
Compound 12 has a molecular formula of C19H31O8P at m/z 417.3([M-H]-),consistent with a structure having one more phosphoryl group than 11.The unique difference between 12 and 11 is C-9 proton signal.It is downfield shifted to δH4.30(1H,m)from δH3.65(1H,m)while the other proton signals of C-8 and C-10 have a slight shifts.So the phosphorylation must occur at C-9.
A molecular formula of C20H32O5for compound 13 was established by the NMR spectrum and HRMS data.A 14 mass units increase than 11 suggests that a methyl group must exists in 13.In the1H NMR spectrum of 13,an obvious proton signalδH3.55(3H,s)was observed showing a methoxyl group must lie in C-1 because of other proton signal keep consistent with its of 11.It was convinced from its13C NMR,in which a shift from δC182.7 upfield shifted to δC175.8 accompanied with the minor change of C-2 carbon signal.
Compound 14 was obtained as a light-yellow oil,and its molecular formula was determined to be C20H33O8P by Q-TOF-HRMS at m/z 431.1834[M-H]-(calculated as 431.1835).It is quite easy to determine the structure of 14.1H and13C NMR analyses(Table 2,3)revealed 14 contains a phosphoryl group at C-9 like 12 and a methoxyl group at C-1 like 13.

Table 1 1H(400 MHz)NMR spectral data of 10 and 11 in MeOD

Table 2 1H(400 MHz)NMR spectral data of 12-14 in MeOD

2 2.16(2H,t,8) 2.07(2H,t,8) 2.16(2H,t,8)3 1.29-1.60(2H,m) 1.29-1.60(2H,m) 1.29-1.60(2H,m)4 1.29-1.60(2H,m) 1.29-1.60(2H,m) 1.29-1.60(2H,m)5 4.01(1H,m) 3.91(1H,dd,7,15.5) 4.01(1H,m)6 5.69(1H,dd,7,17.5) 5.38(1H,dd,7,17.5) 5.69(1H,dd,7,17.5)7 5.51(1H,dd,7,15.5) 5.51(1H,dd,7,15.5) 5.51(1H,dd,7,15.5)8 2.18(1H,m) 2.11(1H,m) 2.18(1H,m)8-CH3 1.13(3H,d,6.8) 0.94(3H,d,6.8) 1.13(3H,d,6.8)9 4.30(1H,m) 3.65(1H,m) 4.30(1H,m)10A 1.29-1.60(2H,m) 1.29-1.60(1H,m) 1.29-1.60(2H,m)10B 1.29-1.60(2H,m) 1.29-1.60(1H,m) 1.29-1.60(2H,m)11 4.99(1H,m) 4.70(1H,m) 4.99(1H,m)12 5.47(1H,t,10) 5.36(1H,t,10) 5.47(1H,t,10)13 6.40(1H,t,11.6) 6.35(1H,t,11.6) 6.40(1H,t,11.6)14 6.26(1H,t,11) 6.07(1H,t,11) 6.26(1H,t,11)15 5.98(1H,t,11) 5.90(1H,t,11) 5.98(1H,t,11)16 6.55(1H,t,11.6) 6.46(1H,t,11.6) 6.55(1H,t,11.6)17 5.71(1H,dd,7) 5.70(1H,dd,7) 5.71(1H,dd,7)18 1.79(3H,d,6.7) 1.70(3H,d,6.7) 1.79(3H,d,6.7)
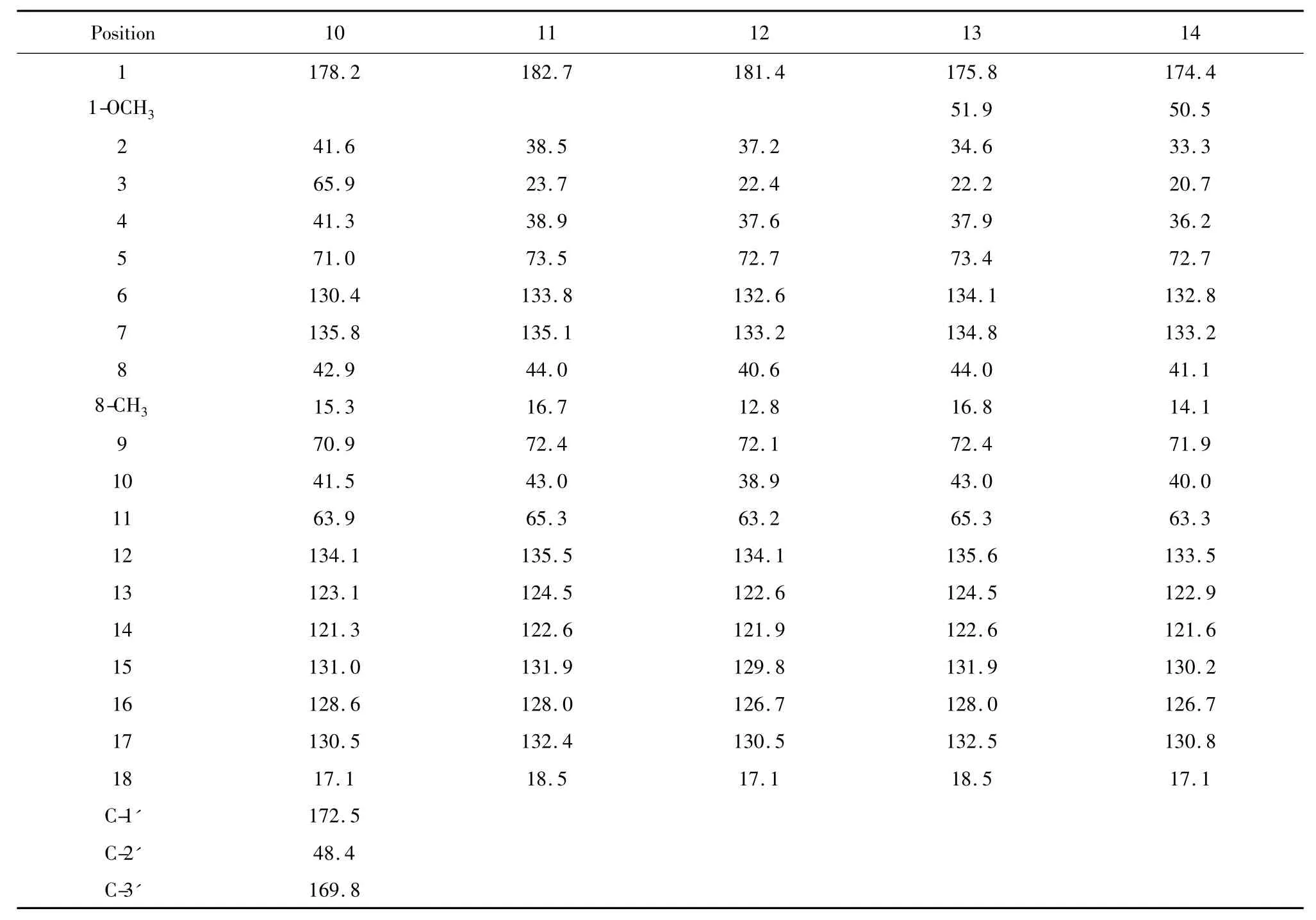
Table 3 13C(100.61 MHz)NMR spectral data of 10-14 in MeOD
Elucidation of their proposed biosynthesis pathway
In the fostriecin biosynthesis,its polyketide chain biosynthesis is obeyed by the strict PKSs biosynthesis routine including PKS chain start,elongation and termination following by the specific post-PKS modification steps.A malonated acid moiety is esterized to C3-OH by a potential enzymes(remaining to be elucidate in the next work)before the chain liberated from PKSs;Then,the PKS chain is cycled to form a six-membered ring to produce compound 4(Figure1).From then on,three hydroxylations and one phosphorylation occur at the compound 4 to produce fostriecin.The isolation of compounds 11-14 brings a new chance to elucidate the PKS biosynthesis mechanism for fostriecin or other polyketides.
Considering their fermentation productivity compounds 10-14 should be derived from the abnormal polyketide biosynthesis or post-PKS modification accompanied by the normal fostriecin biosynthesis.The abnormal or partial TE function leads to the production of 10.The other possibility was it derived from the degradation of 4.However,the isolation of compound 11 brought the huge difficulty to the common polyketide biosynthesis mechanism.
The C-3 hydroxyl group during PKS elongation process is restored to the saturated bond by unknown biosynthesis mechanism in 11.Three proposed pathways were proposed.Firstly,the saturated bond of C2-C3 maybe from the abnormal de-malonic acid reaction or other elimination reaction in vivo as compound 4 to produce fostriecin shown in pathway 1.The routine of PKSs theory could not be used for elucidation.Secondly,a nascent polyketide chain was liberated to produce 15,which could not be achieved and not detected from HRMS analysis to the data.Then,it occurred two tailoring steps to produce 11 with little possibility as listed in pathway 2.Thirdly,as shown in pathway 3,a serial tailoring steps maybe occurs before PKSs chain termination to directly produce 11.We could not exclude the possibility of borrowing DH/ER domain from module 1,which jointly performs the saturated bond formation in the common polyketide biosynthesis.The biosynthesis of 11-14 is produced from the post-PKS modification of 11 by fosH or other enzyme.Although we could not accurately demonstrate the formation reason of saturated bond in PKS which be carried out in the future work,this work still provides a new insight to elucidate novel polyketide biosynthesis mechanism and develop new drug candidate of fostriecin.

Fig.2 Three proposed biosynthesis pathway of compound 11
Acknowledgements This research work was finan- cially supported by National Natural Sciecnce Funds for Distinguished Young Scholar 30688003 to L.T.
1 Staunton J,Weissman KJ.Polyketide biosynthesis:a millennium review.Nat Prod Rep,2001,4:380-416.
2 Fischbach M,Walsh CT.Assembly-Line enzymology for polyketide and nonribosonmal peptide antibiotics:logic,machinery and mechanisms.Chem Rev,2006,8:3468-3496.
3 Tunac JB,Graham BD,Dobson WE.Novel antitumor agents CI-920,PD 113,270 and PD 113,271.I.Taxonomy,fermentation and biological properties.J Antibiot,1983,36:1595-1600.
4 Stampwala SS,Bunge RH,et al.Novel antitumor agents CI-920,PD 113270 and PD 113271 II.Isolation and characterization.J Antibio,1983,36:1601-1605.
5 Lewy DS,Gauss CM,et al.Fostriecin:chemistry and biology.Curr Med Chem,2002,9:2005-2032.
6 Leopold WR,Shillis JL,et al.Anticancer activity of the structurally novel antibiotic CI-920 and its analogues.Cancer Res,1984,44:1928-1932.
7 Boritzki JT,Wolfard ST,Besserer JA.Inhibition of typeⅡtopoisomerase by fostriecin.Biochem Pharmacol,1998,37:4063-4068.
8 Walsh AH,Cheng A,Honkanen RE.Fostriecin,an antitumor antibiotic with inhibitory activity against serine/threonine protein phosphatases types 1(PP1)and 2A(PP2A),is highly selective for PP2A.FEBS Lett,1997,416:230-234.
9 Kong RX,Liu XJ,et al.Elucidation of the biosynthetic gene cluster and the post-PKS modification mechanism for fostriecin in Streptomyces pulveraceus.Chem Biol,2013,20:45-54.
10 Liu XJ,Kong RX,et al.Identification of the post-Polyketide synthase modification enzymes for fostriecin biosynthesis in Streptomyces pulveraceus.J Nat Prod,2013,76:524-529.

