Low Trichorhinophalangeal Syndrome 1 Gene Transcript Levels in Basal-like Breast Cancer Associate with Mesenchymal-to-epithelial Transition△
Yi Bao, Ling-juan Ruan, and Juan-fen Mo
Key Laboratory, Jiaxing Second Hospital, Jiaxing University, Jiaxing, Zhejiang 314000, China
TRICHORHINOPHALANGEAL syndrome 1 (TRPS-1) gene, one of GATA family transcription factors, was initially identified as an essential gene associated with trichorhinophalangeal syndromes, a rare autosomal genetic disease.1The conventional GATA transcription factors have two GATA zinc finger regions, whereas TRPS-1 protein only contains a single GATA zinc finger region.2GATA zinc finger domain of TRPS-1 has 77% conservation with other GATA family members.2Experimental evidences indicate that TRPS-1 identifies and binds with the GATA binding sequence (T/A) GATA (A/G) in the promoter regions of target genes.2
A recent bioinformatics analysis illustrated that TRPS-1 is one of the genes among the identified 22 transcription factors that are most enriched in terminal end buds and in mature ducts of mammary glands.3In other words, TRPS-1 may be associated with luminal progenitor cell differentiation during the development of mammary glands. It has been reported that TRPS-1 is co-expressed with bone morphogenetic protein 7, a molecule playing key roles in mesenchymal-to- epithelial transition that is associated with kidney tubule cells differentiation.4Thus, TRPS-1 mediates epithelial cell differentiation in renal tubule during earlier renal development.4
Breast cancer is characterized into several subtypes on the basis of gene expression profiles. The hormone receptors, estrogen receptor alpha (ERa) and progesterone receptor positive types, are the most predominant molecular subsets that are termed as the luminal A and luminal B subtypes.5Human epidermal growth factor receptor-2 (HER2) overexpressed and basal-like subgroups constitute the major hormone receptor- negative subtypes.5Some research studies have also mentioned other less well-characterized molecular subsets such as normal breast-like and Claudin low.6
BRCA1 plays crucial roles in DNA damage repair, cell cycle control, transcriptional regulation and ubiquitination.7The abnormally short version of the BRCA1 protein due to mutation has been proposed to be a key molecular mechanism in the development of ‘basal-like' or ‘triple negative' breast cancers in young patients.7Exogenous over-expression Slug tends to reduce the expression of BRCA1 in basal-like breast cancers;8further experimental evidence suggested that both Slug and Snail can directly repress BRCA1 expression.9
Recent evidence has pointed out that FOXC1 and CXCL1 serve as molecular markers of basal-like breast cancer. In breast cancer patients, FOXC1 appears to be controlled at the transcriptional level since methylation of the FOXC1 promoter region affects FOXC1 gene expression.10Moreover, FOXC1 also serves as an independent prognostic marker indicating survival.10Exogenous over-expression of FOXC1 induces a progenitor-like phenotype in differentiated mammary epithelial cells.11CXCL1 is another gene that acts as a basal/myoepithelial marker in breast cancer.12Recently, a study reported that CXCL1 is associated with myeloid cell recruitment as it mediates breast cancer metastasis.13
In this study, we proposed to investigate TRPS-1 expression patterns in different subtypes of breast cancer and its correlations with other genes and survival using microarray data sets. We aimed to illuminate whether the expression of TRPS-1 represents a prognostic marker.
MATERIALS AND METHODS
Microarrays
Two published breast cancer microarray data sets—Neth- erlands Cancer Institute (NKI) cohort14and Wang cohort15, were used in this study. NKI data were generated from Affymetrix platform. The gene expression profiling and clinical data of NKI dataset can be downloaded from http://microarray-pubs.stanford.edu/wound_NKI/. In summary, the cohort contains data measured on 25 000 spot oligonucleotide arrays obtained from 295 cases of breast carcinoma. All these patients were lymph-node-negative, younger than 53 years old and stage I or II disease.14Wang data were generated from Agilent platform and measured on the expression of 22 000 transcripts.15In this cohort, 286 cases of breast cancer patients were included. All these patients were lymph-node-negative and had not received adjuvant systemic treatment.15
In practice, luminal A and luminal B subtypes have better prognostic outcome. In contrast, the clinical prognostic outcome of basal-like subtype is poor. In this study, we investigated the expression patterns of TRPS-1 at transcriptional levels in various subtypes of breast cancer by using NKI cohort. In this microarray data set, patients were divided into luminal A, luminal B, basal-like, HER2 overexpressed, and normal breast-like five subtypes.
To explore the expression of TRPS-1 in the association with overall survival (OS) and relapse-free survival (RFS) in breast cancer patients, we analyzed the TRPS-1 expression profiling and clinical data by using NKI cohort. All patients were divided into two groups: one group was TRPS-1 expression in the top 25th percentile and the other group was TRPS-1 expression in the bottom 25th percentile.
Cell culture
MDA-MB-231, a basal-like breast cancer cell line, and MCF-7, an ER+ breast cancer cell line were obtained directly from the American Type Culture Collection (ATCC), and grown in the media containing the following components: DMEM/F12 (Gibco Corporation, California, USA), 10% fetal bovine serum and 1% penicillin/streptomycin (Gibco Corporation). Cells were maintained at 37°C in humidified incubator with 5% CO2.
Western blotting
Cells were lysed in 1xRIPA buffer (Cell Signaling Technology, Massachusetts, USA) with protease inhibitor (Santa Cruz Biotechnology, Texas, USA). Twenty micrograms of total protein lysate were electrophoresed on a polyacrylamide gel, and transferred to 0.45 μm PVDF membrane. The membrane was blocked in TBS-T with 5% non-fat-milk. Anti-TRPS-1 primary antibody (R&D Systems, Minneapolis, USA) was used at 1:1000 in TBS-T for an overnight incubation at 4°C. β-actin immunoblotting was used as a loading control (1:5000; Sigma-Aldrich Inc., Missouri, USA). After incubating with a horseradish peroxidase-conjugated secondary antibody (1:5000; Jackson ImmunoResearch Laboratories, Pennsylvania, USA), signals were detected using an enhanced chemiluminescence kit (GE Healthcare, Little Chalfont, UK).
Statistical analysis
Data were presented as means±SD. Differences in means were analyzed using Student's t test and one-way analysis of variance. For the purpose of studying correlations, Pearson's correlation coefficient was determined by Univariate Cox regression analysis. The Pearson correlation coefficient is a method to measure the correlation (linear dependence) of two variables X and Y, giving a value between +1 and -1. Kaplan-Meier survival curve was used to analyze survival and the P value was calculated by the Log-rank (Mantel-Cox) method. The graph and statistical analysis were performed by GraphPad Prism 5.0 software (GraphPad Software, www.GraphPad.com). P-values<0.05 were considered statistically significant.
RESULTS
Expression pattern of TRPS-1 in different subtypes of breast cancer
There had a significant difference in TRPS-1 transcripts among five subgroups, with the highest expression of TRPS-1 in luminal A subtype but the lowest expression in the basal-like subtype (P<0.001, Fig. 1).
Expression of TRPS-1 protein in breast cancer cell lines
Western blotting showed TRPS-1 signal was nearly undetectable in MDA-MB-231 cells, as compared with MCF-7 cells with strong TRPS-1 expression (Fig. 2).
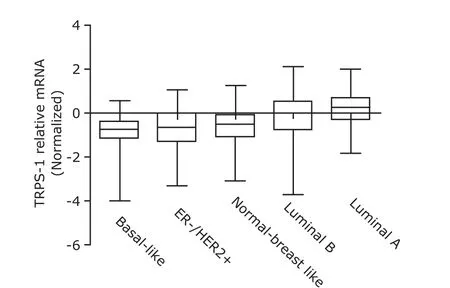
Figure 1. Analysis result of TRPS-1 gene expression in various subtypes of breast cancer by using NKI corhort. TRPS-1: trichorhinophalangeal syndrome 1; NKI: Netherlands Cancer Institute; ER: estrogen receptor; HER2: epidermal growth factor receptor-2. TRPS-1 mRNA levels were lowest in basal-like breast cancer and the difference was significant among five groups (all P<0.0001).
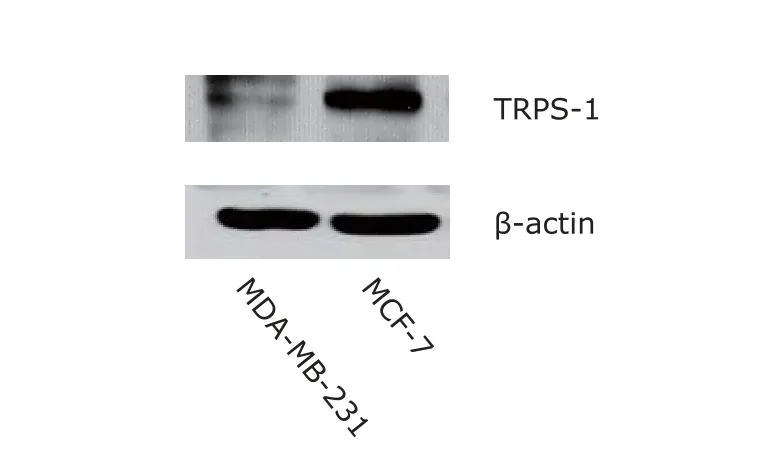
Figure 2. TRPS-1 protein expression in MDA-MB-231 and MCF-7 breast cancer cell lines detected by Western blotting. β-actin was used as a loading control.
Correlation of TRPS-1 with epithelial to mesenchymal transition makers Slug and Snail
In this study, NKI and Wang cohorts, two independent microarray data sets were used to investigate the association between TRPS-1 and other key molecules involving epithelial to mesenchymal transition. A negative correlation was observed between TRPS-1 and Slug, an epithelial to mesenchymal transition maker. Univariate Cox regression analysis showed a negative correlation between TRPS-1 and Slug (P<0.05). The Pearson correlation coefficient (r) was -0.1366 (P=0.0189) and -0.1571 (P=0.0078) in NKI and Wang data set respectively. No significant correlation was observed between TRPS-1 and Snail, another epithelial to mesenchymal transition maker in both data set (r=0.09923, P>0.05, NKI data; r=-0.06562, P>0.05, Wang data).
Correlation of TRPS-1 with genes serving as biomarkers in basal-like breast cancer subtype
As described above, the low TRPS-1 transcripts were presented in basal-like breast cancer subtype. Therefore, we explored the correlation of TRPS-1 with several genes identified as molecular biomarkers of basal-like breast cancer. A significantly inverse correlation was observed between TRPS-1 and FOXC1 by analyzing NKI data set and Wang data set. Analysis of univariate Cox regression demonstrated that all the correlations were statistically significant (P<0.05). The Pearson correlation coefficient (r) was -0.1211 (P=0.0376) and -0.1709 (P=0.0037) in NKI and Wang data set respectively. A negative correlation between TRPS-1 and CXCL1 was also observed. The Pearson correlation coefficient (r) was -0.1197 (P=0.0399) and -0.3436 (P<0.0001) in NKI and Wang data set respectively. A positive correlation between TRPS-1 and BRCA1, a key molecule indicating being dysregulated in early onset of basal-like breast cancer, was also observed. The Pearson correlation coefficient (r) was 0.1728 (P= 0.0029) and 0.1805 (P=0.0022) in NKI and Wang data set respectively.
TRPS-1 might be a novel prognostic factor
To explore the expression of TRPS-1 in the association with OS and RFS in breast cancer patients, we analyzed the TRPS-1 expression profiling and clinical data by using NKI cohort. A statistically significant difference was observed between two groups, and the low TRPS-1 expression was associated with poor OS (hazard ratio 1.79, 95% CI of ratio 0.9894 to 3.238, P=0.054, Fig. 3A) and RFS (hazard ratio 1.913, 95% CI of ratio 1.159 to 3.156, P<0.05, Fig. 3B). P value was calculated using the Log-rank test. The breast cancer patients in the NKI cohort assigned to the poor-prognosis group by the 70-gene signature expressed low TRPS-1 levels (P<0.0001, Fig. 3C). P value was calculated using Student's t test.

Figure 3. Analysis result of overall survival and relapse-free survival of breast cancer patients on the basis of the expression of TRPS-1 by using NKI data set. A. Kaplan-Meier overall survival curves for NKI cohort. B. Kaplan-Meier relapse-free survival curves for NKI cohort. C. TRPS-1 expression in poor and good prognosis groups on the basis of the 70-gene signature. Patients were divided into two groups based on the mRNA levels of TRPS-1. TRPS-1 expression in the top 25th percentile represented one group and TRPS-1 expression in the bottom 25th percentile represented the other.
DISCUSSION
In this study, we analyzed the gene chip data obtained from the public databases. We found that the lowest levels of TRPS-1 transcripts were present in basal-like breast cancer, whereas the highest gene expression was reported in luminal A subtype. This indicates that the loss of TRPS-1 may be one of the characteristics associated with basal-like breast cancer. However, we still cannot decipher how TRPS-1 is regulated in breast cancer. We need also point out that the patients in these databases were recruited from Netherland and the United States. Additionally, they were in early stage and lymph-node-negative. Whether the data obtained here represent Chinese women or not, should be further determined. Moreover, we still need to ascertain whether TRPS-1 plays a pivotal role in ductal endothelial cell differentiation involved in the development of mammary glands. Recently, Stinson and colleagues16reported that miR-221/222 was able to reduce the expression of E-cadherin by regulating 3 ' non-translational regions of TRPS-1 in breast cancer. It has also been reported that TRPS-1 decreases the expression of zinc finger E-box-binding homeobox 2, an E-cadherin transcriptional repressor. This finding serves as an evidence to illustrate that TRPS-1 is probably involved in the suppression of epithelial to mesenchymal transition in breast cancer patients.16These findings are in good agreement with the following hypothesis: the decreased TRPS-1 is to be one of the important molecular phenotypes in basal-like breast cancer. While analyzing the NKI and Wang cohorts, TRPS-1 and Slug had a significantly inverse correlation, but no relevance was observed between TRPS-1 and Snail. This result indicates that TRPS-1 possibly serves as a direct or indirect downstream target of Slug. Snail and Slug have been confirmed as key transcription factors involving epithelial to mesenchymal transition procedure. The expression of these two molecules can be regulated by different pathways. For instance, Slug expression is able to be mediated by the Canonical Wnt signaling pathway;17Snail expression is the downstream of NF-κB in B-raf mutation melanoma patients.18Therefore, TRPS-1 is more likely involving the regulation pathways of Slug. Indeed, a recent research has indicated that knockdown Slug expression using small interfering RNA (siRNA) oligo resulted in a decreased expression of miR-221/222 in the MDA-MB-231 cells.19In this study, authors have reported that miR-221/222 serves as a direct downstream target of Slug when chromatin immunoprecipitation technique is used.19Combined with our findings, current evidences strongly indicate that slug appears as an indirect regulator of TRPS-1.
TRPS-1 is a newly identified GATA family member. GATA3 is another GATA transcription factor that has been studied most extensively. It has been reported that GATA3 plays multiple roles in the maturation of mammary glands and the development of breast cancer. GATA3 protein was detected in the mammary epithelium cells but not in the myoepithelial cells.20In an immunohistochemical study, it was found that TRPS-1 co-expressed with GATA3 in ductal epithelial cells of mammary glands.20In mice, a specific knockout GATA3 gene was reported in the breast tissue, wherein the normal differentiation of duct could be terminated.3This indicates that GATA3 plays a pivotal role in ductal epithelial cell differentiation. In addition to promoting the differentiation in the normal breast tissue, GATA3 is also associated with the incidence of breast cancer.3,21,22High expression of GATA3 protein serves as a good prognostic indicator in breast cancer patients.23There is a positive correlation of TRPS-1, GATA3 and ER in breast cancer,20indicating a close association among these three transcription factors. But the regulating circuits between GATA3 and TRPS-1 need to be further clarified.
We further examined the correlation of TRPS-1 with FOXC1 and CXCL1, two genes reported to be markers of basal-like breast cancer. In this study, we observed that TRPS-1 transcript levels were inversely correlated with FOXC1 and CXCL1. The expression of FOXC1 is associated with the high expression of epithelial to mesenchymal transition markers, such as vimentin, and loss of E-cadherin in breast cancer.24Elevated protein level of FOXC1 has also been reported to be a poor prognostic indicator in breast cancer.25High expression of FOXC1 triggers chemoresistance in basal-like breast cancer.24A recent study was reported that siRNA was used to silence the expression of FOXC1 in MDA-MB-231, thereby leading to an increased expression of E-cadhrin and decreased expression of fibronectin and vimentin,24indicating FOXC1 is one of transcription factors induced epithelial to mesenchymal transition. In additon, a positive correlation of TRPS-1 and BRCA1 was observed in our current study. It has been reported that GATA3 and BRCA1 can be combined together and act on the promoter region of FOXC1, thereby inhibiting FOXC1 gene transcription.24Given that their inhibition effect on FOXC1 transcription is lost when either BRCA1 undergoes gene mutation or GATA3 is subjected to silencing, this may be a key molecular mechanism of basal-like breast cancer.24Taken together, we have shown that TRPS-1 positively correlated with both GATA3 and BRCA1, indicating that TRPS-1 might play its regulatory roles together with GATA3 and BRCA1. However, we need further investigate whether TRPS-1 has the capability to directly form a complex with GATA3 and BRCA1, which mediates the suppression of FOXC1 expression.
Using Kaplan-Meier survival curve, we observed that high mRNA levels of TRPS-1 have good prognosis using the breast cancer microarray data reported by Van de Vijver and colleagues. In addition, we used a 70-gene prognostic signature to discriminate patients with good and poor prognosis from this dataset,14identifying that a high TRPS-1 expression levels correlated with good prognosis. Recently, it has been reported that 70-gene prognostic signature, the intrinsic subtypes, and the recurrence score were strongly concordant to evaluate breast cancer outcome.26In practice, the clinical outcome of breast cancer can be predicted by tumor stage and pathological subtypes. Based on extensive genomic studies, a range of genes have been selected for good prognostic predictors in breast cancer. In present study, our data indicate that the detection of TRPS-1 transcript level may also be used as an independent indicator to predict the prognosis of breast cancer.
1. Momeni P, Glockner G, Schmidt O, et al. Mutations in a new gene, encoding a zinc-finger protein, cause trichorhinophalangeal syndrome type I. Nat Genet 2000; 24: 71- 4.
2. Malik TH, Shoichet SA, Latham P, et al. Transcriptional repression and developmental functions of the atypical vertebrate GATA protein TRPS1. EMBO J 2001; 20: 1715-25.
3. Kouros-Mehr H, Slorach EM, Sternlicht MD, et al. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 2006; 127: 1041-55.
4. Gai Z, Zhou G, Gui T, et al. Trps1 haploinsufficiency promotes renal fibrosis by increasing Arkadia expression. J Am Soc Nephrol 2010; 21: 1468-76.
5. Reis-Filho JS, Pusztai L. Gene expression profiling in breast cancer: Classification, prognostication, and prediction. Lancet 2011; 378: 1812-23.
6. Prat A, Parker JS, Karginova O, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 2010; 12: R68.
7. Buckley NE, Mullan PB. BRCA1—conductor of the breast stem cell orchestra: the role of BRCA1 in mammary gland development and identification of cell of origin of BRCA1 mutant breast cancer. Stem Cell Rev 2012; 8: 982-93.
8. Proia TA, Keller PJ, Gupta PB, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell 2011; 8: 149-63.
9. Wu ZQ, Li XY, Hu CY, et al. Canonical Wnt signaling regulates Slug activity and links epithelial-mesenchymal transition with epigenetic Breast Cancer 1, Early Onset (BRCA1) repression. Proc Natl Acad Sci USA 2012; 109: 16654-9.
10. Dejeux E, Ronneberg JA, Solvang H, et al. DNA methylation profiling in doxorubicin treated primary locally advanced breast tumours identifies novel genes associated with survival and treatment response. Mol Cancer 2010; 9: 68.
11. Bloushtain-Qimron N, Yao J, Snyder EL, et al. Cell type-specific DNA methylation patterns in the human breast. Proc Natl Acad Sci USA 2008; 105: 14076-81.
12. Bierie B, Stover DG, Abel TW, et al. Transforming growth factor-beta regulates mammary carcinoma cell survival and interaction with the adjacent microenvironment. Cancer Res 2008; 68: 1809-19.
13. Acharyya S, Oskarsson T, Vanharanta S, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 2012; 150: 165-78.
14. van't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002; 415: 530-6.
15. Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 2005; 365: 671-9.
16. Stinson S, Lackner MR, Adai AT, et al. miR-221/222 targeting of trichorhinophalangeal 1 (TRPS1) promotes epithelial-to-mesenchymal transition in breast cancer. Sci Signal 2011; 4: pt5.
17. Heuberger J, Birchmeier W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol 2010; 2: a002915.
18. Lin K, Baritaki S, Militello L, Malaponte G, et al. The role of B-RAF mutations in melanoma and the induction of EMT via Dysregulation of the NF-κB/Snail/RKIP/PTEN circuit. Genes Cancer 2010; 1: 409-20.
19. Lambertini E, Lolli A, Vezzali F, et al. Correlation between Slug transcription factor and miR-221 in MDA-MB-231 breast cancer cells. BMC Cancer 2012; 12: 445.
20. Chen JQ, Litton J, Xiao L, et al. Quantitative immunohisto- chemical analysis and prognostic significance of TRPS-1, a new GATA transcription factor family member, in breast cancer. Horm Cancer 2010; 1: 21-33.
21. Kouros-Mehr H, Kim JW, Bechis SK, et al. GATA-3 and the regulation of the mammary luminal cell fate. Curr Opin Cell Biol 2008; 20: 164-70.
22. Albergaria A, Paredes J, Sousa B, et al. Expression of FOXA1 and GATA-3 in breast cancer: the prognostic significance in hormone receptor-negative tumours. Breast Cancer Res 2009; 11: R40.
23. Yoon NK, Maresh EL, Shen D, et al. Higher levels of GATA3 predict better survival in women with breast cancer. Hum Pathol 2010; 41: 1794-801.
24. Tkocz D, Crawford NT, Buckley NE, et al. BRCA1 and GATA3 corepress FOXC1 to inhibit the pathogenesis of basal-like breast cancers. Oncogene 2012; 31: 3667-78.
25. Ray PS, Wang J, Qu Y, et al. FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Res 2010; 70: 3870-6.
26. Fan C, Oh DS, Wessels L, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med 2006; 355: 560-9.
 Chinese Medical Sciences Journal2013年3期
Chinese Medical Sciences Journal2013年3期
- Chinese Medical Sciences Journal的其它文章
- Medical Foreign Bodies in Urinary Bladder: a Case Report
- Role of Sclerostin in the Bone Loss of Postmenopausal Chinese Women with Type 2 Diabetes
- Neurotoxicity and Biomarkers of Lead Exposure: a Review
- Haploidentical Allogeneic Hematopoietic Stem Cell Transplantation for Thymoma-associated Severe Aplastic Anemia:a Case Report△
- CORRECTION
